Figures & data
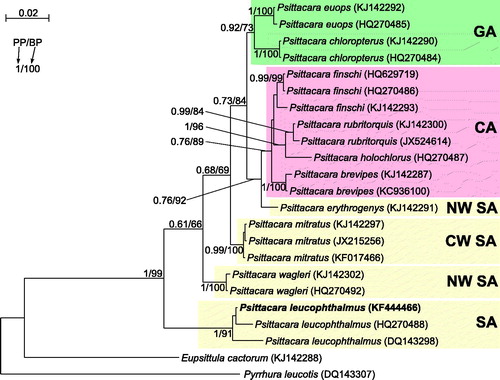
Figure 1. The phylogenetic tree obtained in MrBayes for nd2 gene indicating that the studied individual (bolded) belongs to P. leucophthalmus. The parrot is kept in aviculture and its blood sample from which DNA was isolated is available in the laboratory at the Department of Genetics in Wroclaw University of Environmental and Life Sciences under the number PL16966. Clades with taxa inhabited different geographic regions were marked by various colours/shading: CA – Central America; GA – Greater Antilles; NW SA – northwest South America; CW SA – central-western South America; SA – a vast part of South America. Migration and colonization routes of Psittacara parrots are not easy to infer because related clades include taxa, which currently have distant and restricted distributions. P. erythrogenys is the only South American species that is placed between Central American (finschi, rubritorquis, brevipes, holochlora) and Greater Antillean (euops, chloropterus) taxa. Similarly, P. wagleri with the most northern distribution in South America is placed between mitratus from the central South America, and leucophthalmus widespread in the large part of the continent. The close relationship of north-western P. erythrogenys and the Central American parrots suggests migrations through the Isthmus of Panama. However, origin of parrots from Greater Antilles remains unsolved. Values at nodes, in the order shown, indicate posterior probabilities found in MrBayes (PP) and bootstrap percentages calculated in TreeFinder (BP). In the MrBayes (Ronquist et al. Citation2012) analysis, separate mixed substitution models were assumed for three codon positions with information about heterogeneity rate across sites as proposed by PartitionFinder (Lanfear et al. Citation2012). We applied two independent runs, each using eight Markov chains. Trees were sampled every 100 generations for 10,000,000 generations. After obtaining the convergence, trees from the last 3,938,000 generations were collected to compute the posterior consensus. In the case of TreeFinder (Jobb et al. Citation2004), the separate substitution models were selected for three codon positions according to Propose Model module in this program, and 1000 replicates were assumed in the bootstrap analysis. The posterior probabilities <0.5 and bootstrap percentages <50 were omitted.