Figures & data
Figure 1. Photographs of B. racemose plants showing a typical branching (upper panel) and floral structure (lower panel). Images were taken using a digital camera at Wolmyeong Park (Gunsan, Jeollabuk-do Province, South Korea). Scale bars represent 1 cm.

Figure 2. Schematic map of B. racemosa complete chloroplast genome constructed by CPGview (http://www.1kmpg.cn/cpgview/). The map contains six tracks. From the center to outward, the first track shows the dispersed repeats. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
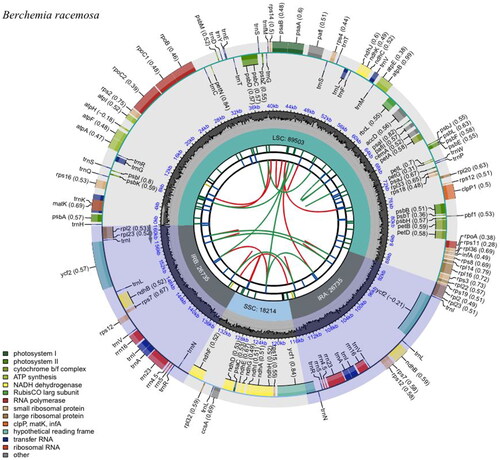
Figure 3. Schematic map of the trans-splicing gene rps12 (A) and cis-splicing genes (B) in the chloroplast genome of B. racemosa. The exons are shown in black; the introns are shown in white. The arrow indicates the sense direction of the gene. The map was generated using CPGview software (http://www.1kmpg.cn/cpgview/).
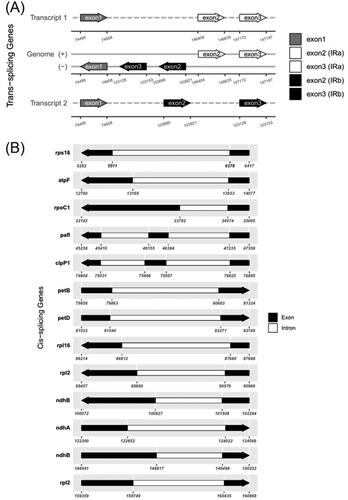
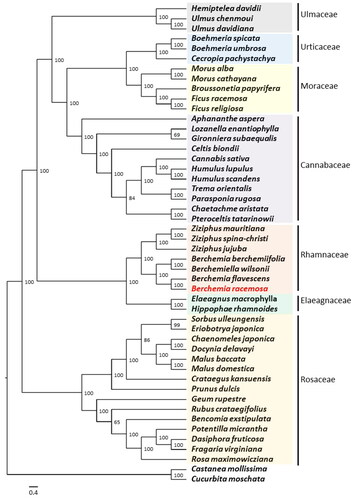
Figure 4. Maximum-likelihood (ML) tree showing the relationship among B. racemosa and representative species within Order Rosales based on the complete chloroplast genome sequences. The following sequences were used: Aphananthe aspera (NC_039726; Zhang et al. Citation2018), Bencomia exstipulata (NC_039924), Berchemia berchemiifolia (NC_037477; Cheon et al. Citation2018), Berchemia flavescens (MK460212; Zhu et al. Citation2019), Berchemia racemosa (ON749761; this study), Berchemiella wilsonii (KY926621; Wang et al. Citation2018), Boehmeria spicata (NC_036989; Huang et al. Citation2019), Boehmeria umbrosa (NC_036990; Huang et al. Citation2019), Broussonetia papyrifera (KX828844), Cannabis sativa (KR363961; Oh et al. Citation2016), Cecropia pachystachya (NC_039763; Wu et al. Citation2017), Celtis biondii (NC_039727; Zhang et al. Citation2018), Chaenomeles japonica (KT932966), Chaetachme aristata (MH118120; Zhang et al. Citation2018), Crataegus kansuensis (MF784433; Zhang et al. Citation2020), Dasiphora fruticosa (MF683841; Zhao et al. Citation2018), Docynia delavayi (KX499860; Zhang et al. Citation2017), Elaeagnus macrophylla (KP211788; Choi et al. Citation2015), Eriobotrya japonica (KT633951; Shen et al. Citation2016), Fragaria virginiana (KY085911), Ficus racemosa (KT368151), Ficus religiosa (KY416513; Bruun-Lund et al. Citation2017), Geum rupestre (NC_037392; Duan et al. Citation2018), Hemiptelea davidii (MK070168; Liu et al. Citation2019), Hippophae rhamnoides (NC_035548; Chen and Zhang Citation2017), Humulus lupulus (KT266264; Vergara et al. Citation2016), Humulus scandens (NC_039730; Zhang et al. Citation2018), Gironniera subaequalis (MH118121; Zhang et al. Citation2018), Lozanella enantiophylla (MH118123; Zhang et al. Citation2018), Malus baccata (KX499859; Zhang et al. Citation2017), Malus domestica (KY818915), Morus alba (KU981119), Morus cathayana (MW465956), Parasponia rugosa (NC_039732; Zhang et al. Citation2018), Potentilla micrantha (HG931056; Ferrarini et al. Citation2013), Prunus dulcis (NC_034696), Pteroceltis tatarinowii (NC_039733; Zhang et al. Citation2018), Rosa maximowicziana (MG727865; Jeon and Kim Citation2019), Rubus crataegifolius (NC_039704; Yang et al. Citation2017), Sorbus ulleungensis (NC_037022), Trema orientalis (NC_039734; Zhang et al. Citation2018), Ulmus davidiana (NC_032718), Ulmus chenmoui (MG581403; Zhang et al. Citation2019), Ziziphus jujuba (NC_030299; Ma et al. Citation2017), Ziziphus mauritiana (NC_037151), and Ziziphus spina-christi (KY628305). Castanea mollissima (KY951992) and Cucurbita moschata (NC_036506) were included as outgroups. The numbers on the nodes indicate bootstrap values from 1000 replicates. The scale bar represents the number of substitutions per site.
Supplemental Material
Download MS Word (116.6 KB)Data availability statement
The genome sequence data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov) under the accession no. ON749761. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR21615884, and SAMN30910605, respectively.
