Figures & data
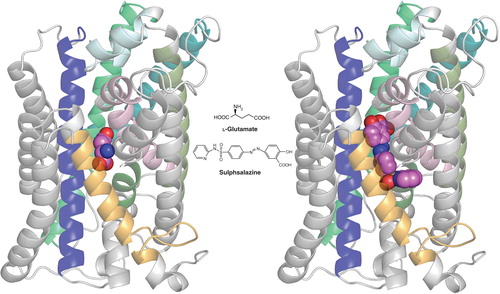
Figure 1. Homology model of xCT with docked ligands. The model was established using the crystal structure coordinates of the amino acid, polyamine and organocation transporter (ApcT) from the RCSB protein databank (3GIA). Protein sequences of human xCT and ApcT were aligned using ClustalW after which the sequences were threaded and the pictured homology model of xCT generated using the default automodel process of MODELLER v9.9. Left shows the xCT model with glutamate docked, right shows it with sulfasalazine docked. The ligand (hot pink) structures were energy minimized using Tripos Sybyl (MMFF forcefield) and docked into the xCT homology model using CCDC GOLD and default settings (GoldScore) system. Modeling was carried out in the University of Montana Molecular Computational Core Facility, in collaboration with S. Patel, N. Natale, M. Braden and J. Gerdes. TMDs 2, 4, 6, 9, 11 and 12 of xCT have either been removed or colored light grey to permit a better view of ligand docking. TMDs shown in color include TMI (light pink), TM3 (dark blue), TM5 (teal), TM7 (olive), TM8 (bright green) and TM10 (gold). Note that only xCT is shown, as it is the subunit responsible for substrate transport. CD98, the regulatory subunit is required for membrane association and is not illustrated.