Figures & data
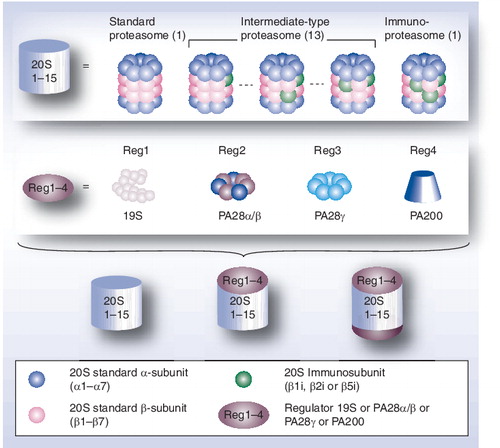
Mammalian proteasome complexes result from the assembly of a 20S core complex, the 20S proteasome, with one or two regulatory particles (RPs) of identical or different protein composition. The 20S core complex is a stable entity assembled in four stacked ring-shape heptamers, α7β7β7α7. Three subunits, β1, β2 and β5, of each β ring exhibit three distinct catalytic activities. These standard β subunits can be completely or partially replaced by three immunosubunits (β1i, β2i and β5i, respectively), which leads to various forms of 20S proteasomes (immunoproteasome and intermediate-type proteasomes). The two α-rings are implicated in the recruitment of RPs at each side of the 20S core complex. In mammals, four activators have been identified: the 19S RPs, PA28αβ, PA28γ and PA200.
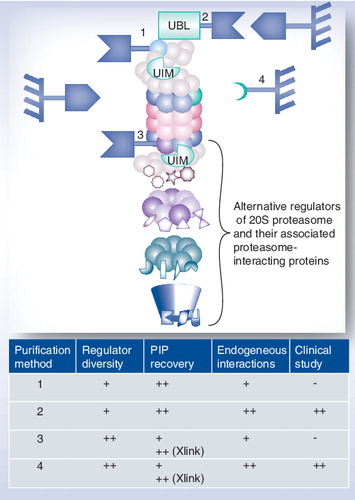
(1) Purification using a tagged 19S regulator subunit, (2) purification using a tagged domain recognizing a subunit of the 19S regulator, (3) purification using a tagged 20S core particle subunit and (4) purification using an antibody specifically directed against a conserved 20S proteasome subunit.
-: Not possible; +: Good; ++: Very good.
PIP: Proteasome-interacting proteins; UBL: Ubiquitin-like; UIM: Ubiquitin-interacting motif; Xlink: Proteins have been in vivo crosslinked prior to purification.
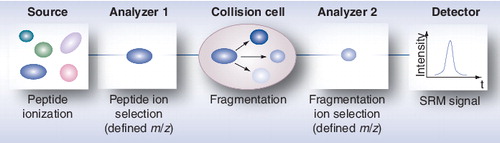
m/z: Mass-to-charge ratio; SRM: Selected reaction monitoring.