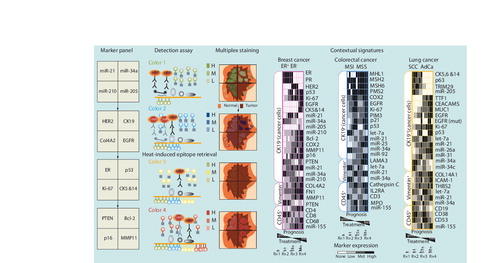
Figures & data
Table 1. Diagnostic and prognostic applications of miRNA signatures in solid tumors.
Table 2. Diagnostic and prognostic applications of single miRNAs in solid tumors.