Figures & data
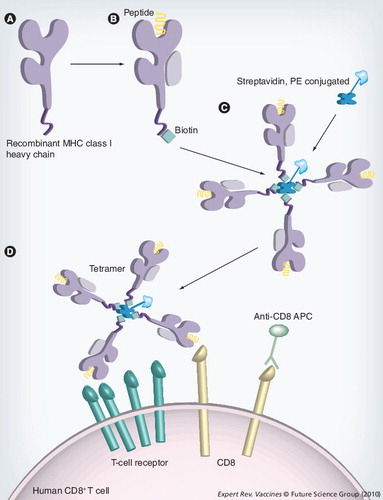
(A) Soluble versions of the heavy-chain MHC class I molecules are synthesized in Escherichia coli. (B) The heavy chain is refolded with β2-microglobulin and the epitope of interest. This refolded MHC molecule is biotinylated with the enzyme BirA at the specific BirA-recognition sequence, at the carboxyl terminus of the MHC molecule. (C)A tetramer is constructed by the addition of four MHC–biotin complexes, binding to a single streptavidin molecule, using the biotin–avidin interaction. This streptavidin molecule is conjugated to a fluorochrome to allow analysis by fluorescence-activated cell sorting. (D) Tetramers are mixed with the cell population that is to be analyzed. The tetramer will bind to CD8+ T cells that are expressing a T-cell receptor that is capable of recognizing the specific peptide and, with the addition monoclonal antibodies, cell populations can then be identified.
APC: Allophycocyanin; PE: Phycoerythrin.
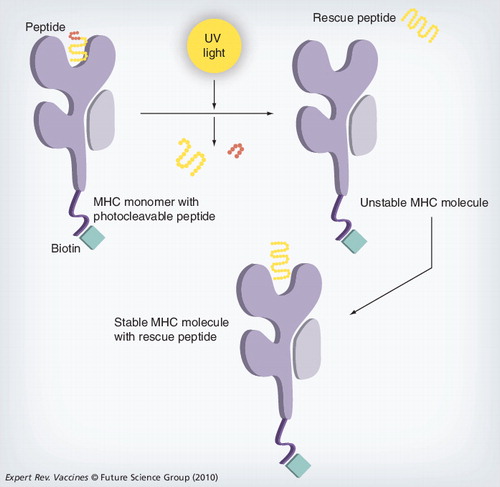
The MHC complex containing the photoliable peptide is irradiated with UV light, cleaving the peptide and producing two fragments that dissociate from the MHC-binding groove. The resulting empty MHC complex is unstable; incubating this MHC with a peptide of interest creates a stable form that can then be used for analysis.
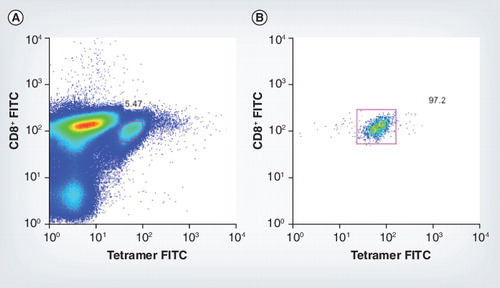
(A) Tetramer-positive mice CD8+ cells for murine cytomegalovirus epitope m45 7 days postinfection, pre-sort. (B) Fluorescence-activated cell sorting plot shows the purity of these tetramer-positive cells post-sort.
FITC: Fluorescein isothiocyanate.
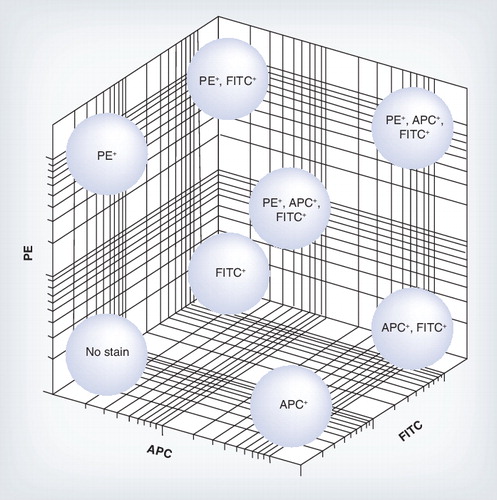
Each tetramer was made by tetramerizing the MHC class I molecule with either one fluorochrome APC, PE and FITC or a mixture, allowing many epitope-specific cell populations to be analyzed from a limited number of fluorochromes.
APC: Allophycocyanin; FITC: Fluorescein isothiocyanate; PE: Phycoerythrin.