Figures & data
Figure 1 Chemical structures of some reported IKKβ inhibitors.
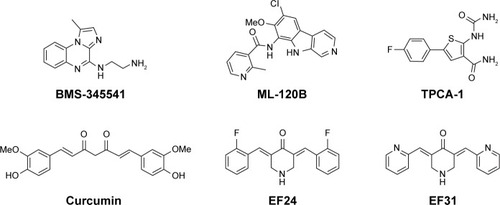
Figure 3 Inhibition of IKKβ kinase by the (A–D) series of analogs of EF24 and EF31 at a concentration of 20 μM was screened through a Caliper mobility shift assay.
Abbreviations: IKKβ, inhibitory kappa B kinase β; IR, inhibitory ratio; Me, methyl; OMe, methoxy.
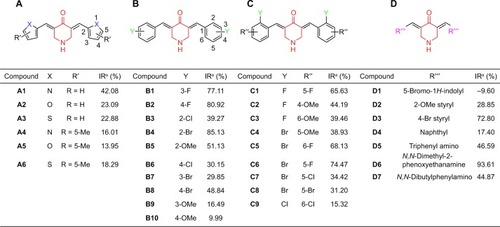
Figure 4 Direct-binding affinity between D6 and IKKβ demonstrated by SPR. Note: Ka, Kd, equilibrium dissociation constant.
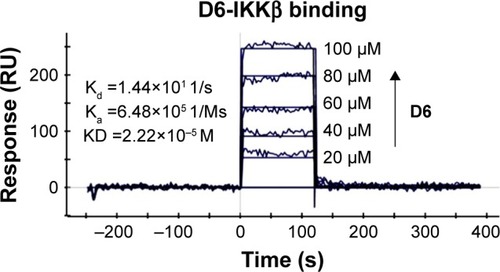
Figure 5 Molecular docking and MD simulation analysis of binding of D6 to the activity cavity of IKKβ.
Abbreviations: IKKβ, inhibitory kappa B kinase β; MD, molecular dynamics; MM-GBSA, molecular mechanics/generalized Born surface area; RMSD, root-mean-square deviation.
Table 1 Six compounds (B1, B2, B4, C6, D3, D6, and CUR) were selected for determining the IC50 values of IKKβ kinase activity, and the active compounds B1, B2, B4, and D6 (IKKβ IC50 <10 μM) were further measured for the antiproliferative effects in three PC cell lines (PANC-1, MiaPaCa-2, and BxPC-3) by the well-established MTT assay
Figure 6 Compound D6 suppressed TNF-α induced phosphorylation of IKKβ and degradation of the downstream signal molecule IκB in a dose-dependent manner.
Abbreviations: DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IKKβ, inhibitory kappa B kinase β; pIKKβ, phosphorylated IKKβ; SEM, standard error of the mean; TNF, tumor necrosis factor.
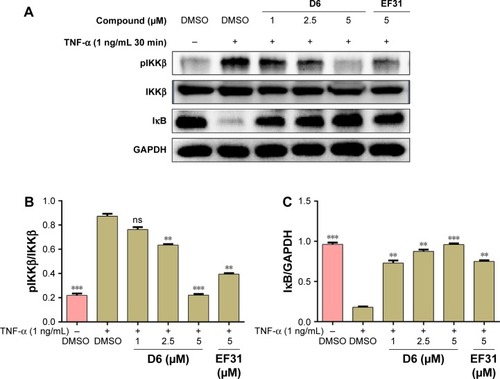
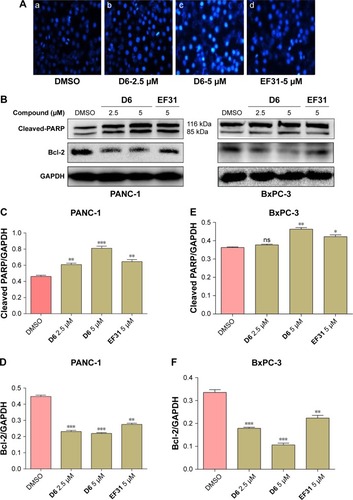
Figure 7 Compound D6 induced PC cell apoptosis.
Abbreviations: DMSO, dimethyl sulfoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; SD, standard deviation.