Figures & data
Figure 1 Domain organization schematic for the NS5A protein.
Notes: (A) D-I (aa 1–213), D-II (aa 250–342), and D-III (aa 356–447) are depicted with green, blue, and red boxes, respectively. Each domain is separated by an LCS (LCS-I and LCS-II). The N-terminal AH is depicted with a yellow box. D1a and 1bCitation33 are designated by dashed green lines. Domain numbering is from Tellinghuisen et al.Citation29 (B) Rows (1–5) show the sequence alignments and domain outlining of the HCV NS5A genotypes 1a, 1b, 2a, 3a, and 4a. Conserved and consensus residues are in red and black on the top line. The critical N-terminal, membrane binding, AH subdomain (aa 1–25) of D1 is outlined in gold. The critical conserved Cys residues that bind Zn++ in subdomain D1a are shaded blue. The proposed RNA-binding subdomain is D1b. Positions 28, 30, 31, and 93 (shaded red) are clinically observed mutations to NS5a-directed inhibitor treatment, which are also associated with high cell-based resistance. Rows (6–8) show the sequence of experimental 3D structures R7G.pdb (by NMR, aa 1–31), 3FQQ.pdb (by X-ray diffraction, aa 31–207), and 1ZH1.pdb (by X-ray diffraction, aa 25–198) based on genotypes 1a, 1b, and 1b, respectively.
Abbreviations: 3D, three-dimensional; aa, amino acid; AH, amphipathic helix; Cys, cysteine; HCV, hepatitis C virus; LCS, low complexity sequence; NMR, nuclear magnetic resonance; NS5A, nonstructural protein 5A; Pdb, protein data bank; RNA, ribonucleic acid; D, domain.
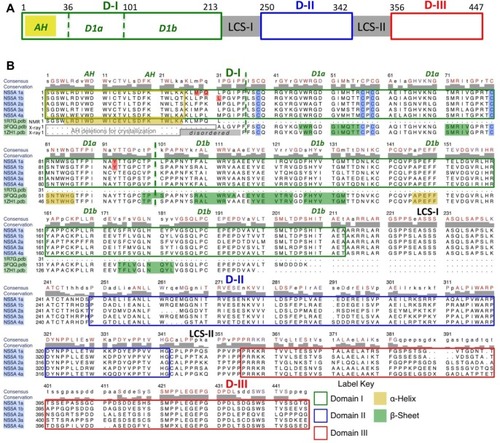
Figure 2 Structure-based theoretical model of membrane-bound NS5A D1 homodimer binding RNA duplex.
Notes: The ribbon representations of the dimeric subunits are colored gold and blue. The N-terminal AHs align in plane with the membrane and pack against residues Y93 and L31 at the dimer interfaces. This AH conformation suggests potential binding site(s) for endogenous cofactors and drugs at the Y93/AH/dimer interfaces that may help explain the activity of NS5A direct inhibitors at different stages of replication and inhibition, and the effects on membranous web morphology. An atom of Zn++ (cyan) binds at the D1a site at the core of each monomer, stabilizing a novel fold. RNA is shown binding at the D1b interface of this dimeric form. The recently identified regions for PI4K-binding at the C-terminal are colored green (182–198), and critical TIP47-binding residues near the AH N-termini are colored magenta. (The templates used PDB ID IZH1, PDB ID IR7G, POPC phospholipid bilayer). Reproduced with permission of James H Nettles. Copyright © 2013.
Abbreviations: AH, amphipathic helix; D1, domain 1 subdomain 1; NS5A, nonstructural protein 5A; PI4K, phosphatidylinositol 4-kinase; PIP4, phosphatidylinositol phosphate 4; pdb, protein data bank; RNA, ribonucleic acid; TIP47.
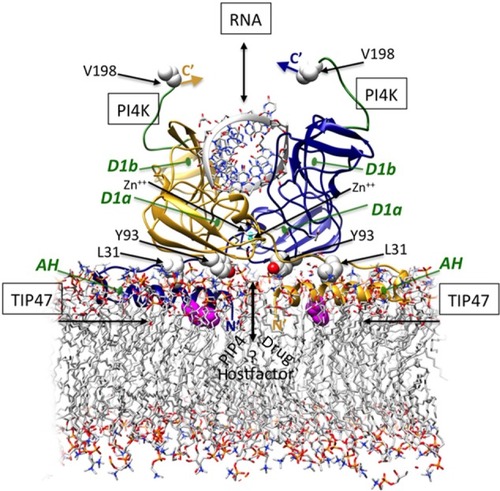
Table 1 Pharmacokinetic parameters of NS5A inhibitors
Figure 3 Representatives of the two general classes of NS5A-directed compounds found in the patent literature.
Notes: The dimeric class 1 compounds (1) (daclatasvir), (2), and (3) (ledipasvir) are the only three currently under clinical evaluation for which structures have been released. The class 2 compounds are diverse chemotypes and include monomers that do not fit class 1 (4–6).
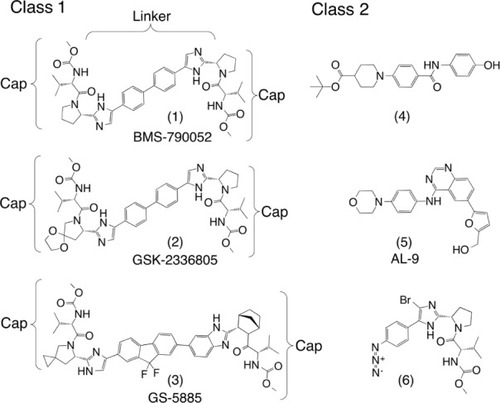
Figure 4 Three chemical series exploring systematic substitutions of a common compound 1 monomeric “cap” and relating specific features associated with activity across both class 1 (dimer) and class 2 (monomer) NS5a directed inhibitors.
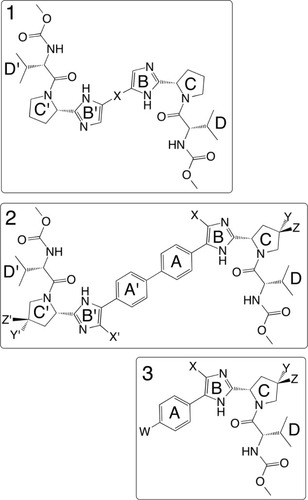
Table 2 Resistance profile and fitness of daclatasvir, in genotype 1a and 1b replicon systemsCitation97
Figure 5 Theoretical models of daclatasvir (turquoise) binding to NS5A and potential AH mediated effect on membrane morphology.
Notes: The models suggest the drug (turquoise) binds simultaneously to two asymmetric sites at the NS5A A/B dimer/membrane interface (blue/gold). A core site between Y93 of each monomer binds one cap of the drug, while the second cap fits between residues 93 and 31 of the aligned subunits. (A) The aromatic linker provides favorable interactions and positions the caps simultaneously within the two LA sites formed by AH aligned in the membrane plane. (B) Conformational change of AH exposes an HA “open” drug site between 93 and 31 of the different subunits. Binding to this state may lock NS5A into a conformation conducive to lipid droplet formation and release to cytosol, and is thought to impair assembly of other viral oligomers. (C) NS5A binding with RNA, NS5B, and other proteins induces replication complex formation in a membranous web and significantly lowers the affinity for drug binding. Reproduced with permission of James H Nettles. Copyright © 2013.
Abbreviations: AH, amphipathic helix; HA, higher affinity; LA, lower affinity; Leu, leucine; NS5A, nonstructural protein 5A; RNA, ribonucleic acid; Tyr, tyrosine.
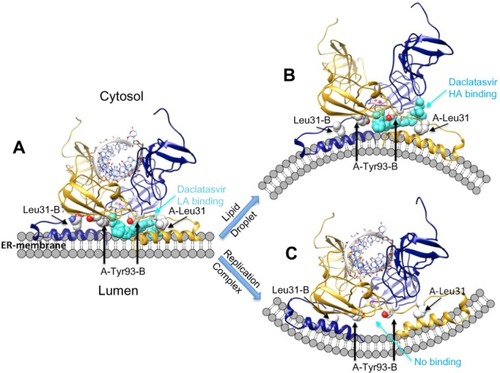
Table 3 NS5A agents in clinical development for chronic HCV infection
