Figures & data
Figure 1 Pharmacophore fingerprints.
Notes: A pharmacophore fingerprint is the representation of a small molecule ligand (A) annotated with molecular interaction features (B) into a string. Typically, every possible three- (or four-) point combination of molecular interaction features (C), with different distances between the features, calculated either through space or by the number of bond lengths (D), is calculated and the frequency of occurrence is stored in a string (E). Such strings are useful for the easy comparison of similarity between multiple molecules.
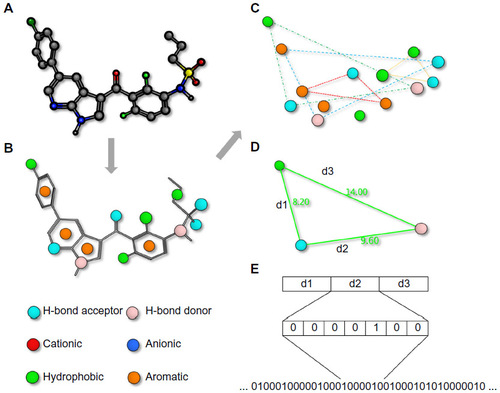
Figure 2 Pharmacophore query.
Notes: A pharmacophore query is comprised of different features. The features represent molecular recognition motifs such as hydrogen bond acceptors or donors, anionic, cationic, hydrophobic, and aromatic groups. The radius of the sphere determines the strictness of the geometric constraint. For features where the correct orientation of the interaction is important such as hydrogen bonds and the aromatic plane, a second feature can be used indicating the vector of the interaction (or the normal of the plane). A pharmacophore query can combine any of these features, with different radii and logic operations such as “AND,” “OR,” and “NOT.” On the left a hypothetical pharmacophore query for BRAF kinase is given.
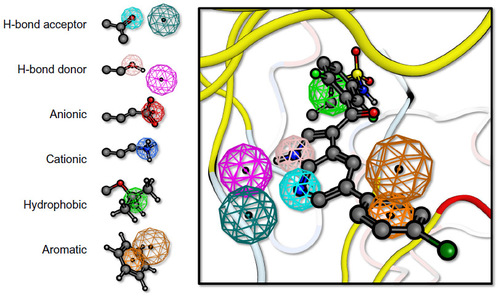
Figure 3 Four different situations for the pharmacophore search.
Notes: The figure shows the four different situations that may be encountered when starting a virtual screening. The situations include the absence of both the ligand and protein structure information, where except for divination, experimental screening is the only option. The second option is the presence of active ligands, but the protein structure is unknown, where pharmacophores can be used for ligand-based virtual screening. The best situation is when binding ligand and structural information is present. The most challenging option is when only a protein structure is available.