Figures & data
Figure 1 Expression and transactivation of dCas9.nVP64.
Abbreviations: CMV, cytomegolovirus; eGFP, enhanced green fluorescent protein; HA, hemagglutinin; NLS, nuclear localization sequence; n.s., not significant; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element.
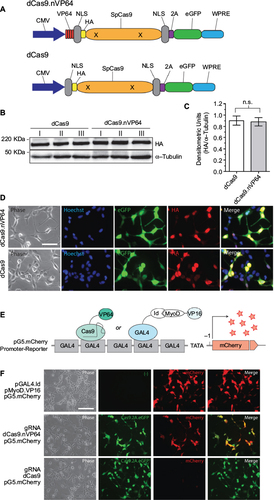
Figure 2 Loci-specific transactivation of the mouse TIMP promoters using dCas9.nVP64.
Abbreviation: TIMP, tissue inhibitor of metalloproteinases.
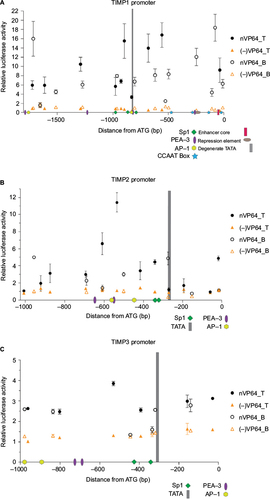
Figure 3 Comparison of the transactivation efficiency of TIMP1 and TIMP2 promoters using different transcriptional activating effectors.
Abbreviations: CMV, cytomegolovirus; eGFP, enhanced green fluorescent protein; HA, hemagglutinin; NLS, nuclear localization sequence; SAM, synthetic activation motif; TIMP, tissue inhibitor of metalloproteinases; VPR, VP64-p65-Rta; WPRE, Woodchuck hepatitis virus post-transcriptional regulatory element.
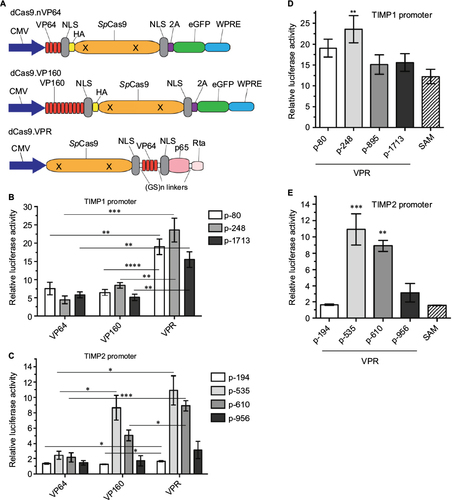
Figure 4 Endogenous TIMP gene activation in mouse NSC34 cells.
Abbreviations: DMSO, dimethylsulfoxide; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; n.s., not significant; PMA, phorbol-12-myristate-13-acetate; TIMP, tissue inhibitor of metalloproteinases; VPR, VP64-p65-Rta.
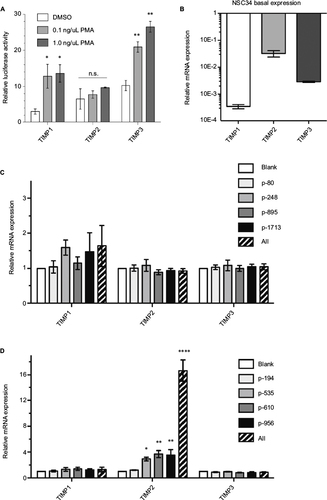
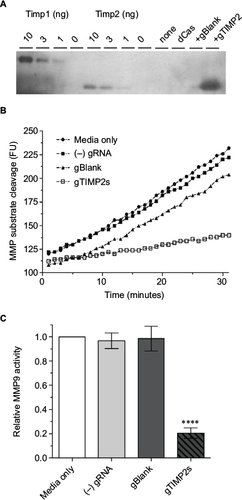
Figure 5 Production of functional TIMP2 protein by gRNA-induced NSC34 cells.
Abbreviations: MMP, matrix metalloproteinase; TIMP, tissue inhibitor of metalloproteinases; VPR, VP64-p65-Rta.