Figures & data
Table I. The top 40 genes in the cluster Up-I.
Table II. The top 20 genes in the cluster Down-I.
Figure 1. The effects of heat on cell viability and the cell cycle in two NHF cell lines, Hs68 and OUMS-36. Cells were exposed to MHT at 41°C for 30 min or HT at 44°C for 30 min. After heat treatment, the cells were incubated for 24 h at 37°C. The cell viability (A, B) and cell cycle (C, D) were measured. (A, C) Hs68 cells. (B, D) OUMS-36 cells. Data indicate the means ± SD for four different experiments. *P < 0.05 versus the control (37°C treatment).
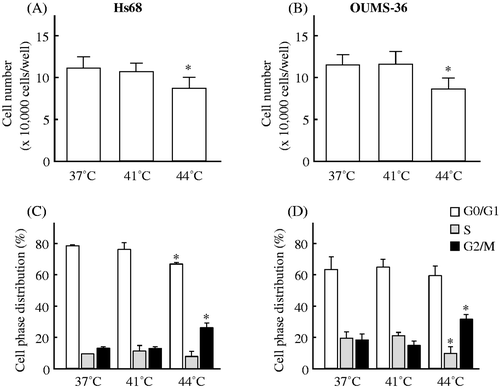
Figure 2. Gene expression analysis. Gene expression analysis of the probe sets that were differentially expressed by a factor of 1.5 or greater in Hs68 and OUMS-36 cells treated with MHT at 41°C for 30 min was performed using bioinformatics analysis tools. (A) Venn diagram of the probe sets that were differentially expressed in Hs68 and OUMS-36 cells. The numbers of probe sets are shown. (B) K-means clustering of commonly and differentially expressed probe sets (1,196 probe sets). Probe sets were grouped in three clusters with distinct expression profiles, Up-I, Down-I and Down-II. The numbers of commonly expressed probe sets and genes are shown. C, control.
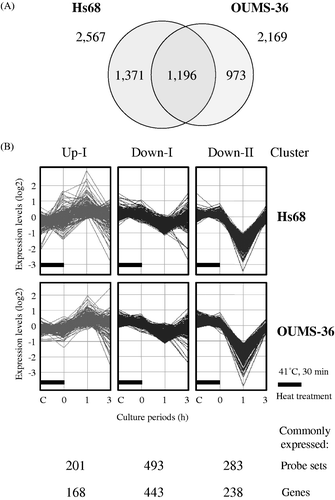
Table III. The top 20 of genes in the cluster Down-II.
Figure 3. Gene networks. Up-regulated genes in the cluster Up-I were analysed by Ingenuity® Pathways Analysis tools. Gene networks E (A) and S (B) associated with ER stress and cell survival, respectively, were identified. The network is displayed graphically as nodes (genes) and edges (the biological relationships between the nodes). The node colour of genes indicates the expression level of genes. Nodes and edges are displayed using various shapes and labels that present the functional class of genes and the nature of the relationship between the nodes, respectively.
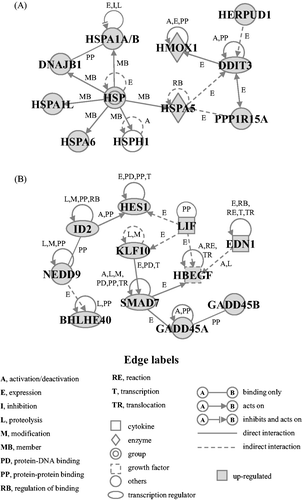
Figure 4. Effects of MHT on the time-course of changes in the expression levels of genes in the gene network E. Cells of the NHF lines Hs68 (Hs), KD, NTI-4 (NTI) and OUMS-36 (OUMS) were incubated at 41°C for 30 min and then cultured at 37°C for 0, 1 and 3 h. Real-time qPCR was carried out. DDIT3 (A), DNAJB1 (B), HERPUD1 (C), HMOX1 (D), HSPA1A/B (E), HSPA1L (F), HSPA5 (G), HSPA6 (H), HSPH1 (I) and PPP1R15A (J) were included in the gene network E. The expression level of each mRNA was normalised to the GAPDH expression level. Data are presented as the mean ± SD (n = 4). *P < 0.05 versus the control.
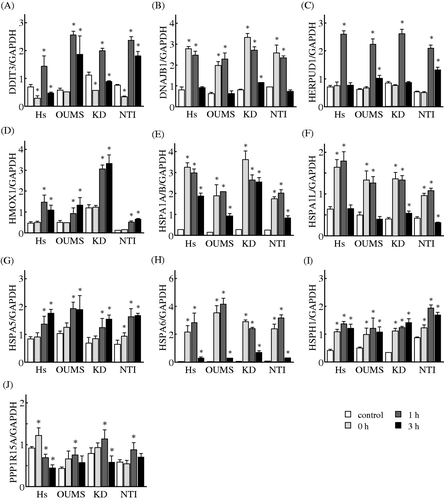
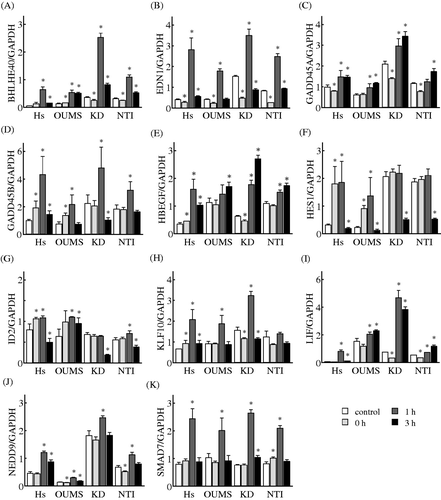
Figure 5. Effects of MHT on the time-course of changes in the expression levels of genes in the gene network S. Cells of the NHF cells Hs68 (Hs), KD, NTI-4 (NTI) and OUMS-36 (OUMS) were incubated at 41°C for 30 min and then cultured at 37°C for 0, 1 and 3 h. Real-time qPCR was carried out. BHLHE40 (A), EDN1 (B), GADD45A (C), GADD45B (D), HBEGF (E), HES1 (F), ID2 (G), KLF10 (H), LIF (I), NEDD9 (J) and SMAD7 (K) were included in the gene network S. The expression level of each mRNA was normalised to the GAPDH expression level. Data are presented as the mean ± SD (n = 4). *P < 0.05 versus the control.