Figures & data
Figure 1. An example of a 2D digital photo of the macroscopic intersection (a) and the corresponding 2D sagittal MR image (b) with delineated structures [cervix-uterus (blue), uterine cavity (green), GTV (red)]. The cervix-uterus structures were segmented from the background on the photo (c) and from surrounding tissue on MRI (d).
![Figure 1. An example of a 2D digital photo of the macroscopic intersection (a) and the corresponding 2D sagittal MR image (b) with delineated structures [cervix-uterus (blue), uterine cavity (green), GTV (red)]. The cervix-uterus structures were segmented from the background on the photo (c) and from surrounding tissue on MRI (d).](/cms/asset/081c4cb6-4a85-4330-8794-d3f83f4a2c04/ionc_a_983655_f0001_oc.jpg)
Figure 2. Box-and-whisker plots of the DSC and the SDE between the MR image and the pathology photo (a, b) and between adjacent MR images and the pathology photo (c, d) after each step in the three-step multi-image registration strategy (rigid registration, affine registration and deformable registration) for the cervix-uterus and the uterine cavity structure. The box represents the upper and lower quartiles (IQR) and the band (red) inside the box the median value. The whisker represents the highest (lowest) value within 1.5 IQR of the upper (lower) quartile. Dots above or below the whiskers are considered outliers. Horizontal lines indicate statistical significant difference (p < 0.01).
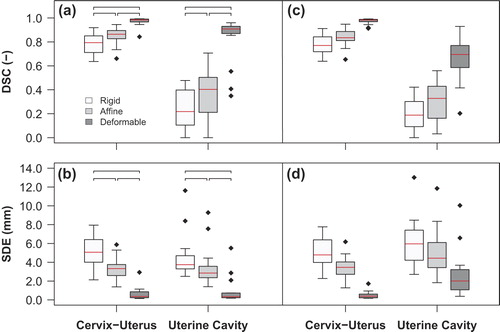
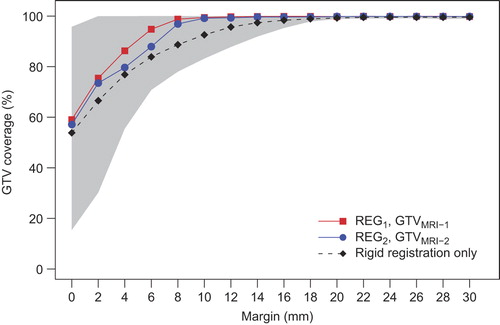
Figure 3. Median GTV coverage after three-step multi-image registration based on GTVMRI-1 after REG1 (red squares) and based on GTVMRI-2 after REG2 (blue circles) as a function of applied margin to the GTVMRI within the cervix-uterus structure. The black dashed line shows the average median GTV coverage of both observations after only rigid registration and the gray region indicates the 95% confidence interval of both observations after three-step multi-image registration.