Figures & data
Figure 1. The transforming growth factor β (TGFβ) pathway. TGFβ ligands function as dimers and signal through type I and type II serine/threonine kinase receptors. Upon ligand binding, the receptors form heterotetrameric complexes allowing the type II receptors to phosphorylate and activate the type I receptors. Subsequently, receptor-activated Smads (R-Smads) are recruited to the type I receptors and phosphorylated. The activated R-Smads associate with the Co-Smad, Smad4, and this heteromeric complex translocates to the nucleus to participate in the transcriptional control of specific target genes (Citation94). TGFβ can also activate various non-Smad signaling pathways.
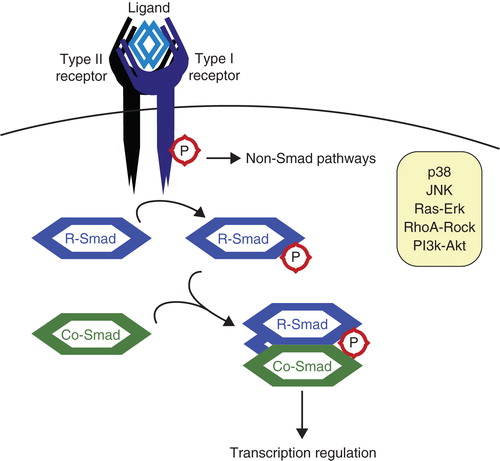
Figure 2. The ubiquitin system. A: Free ubiquitin is bound by the active cysteine residue of an E1 ubiquitin-activating enzyme, and this process requires ATP. Next, Ub is transferred onto the active cysteine of an E2 ubiquitin-conjugating enzyme. Finally, Ub is transferred onto a lysine residue of the target protein by an E3 ubiquitin ligase. Ubiquitin can be conjugated as mono-ubiquitin (B) or in chains such as K63 chains (C) or K48 chains (D), the last-mentioned known for targeting substrates for proteasomal degradation.
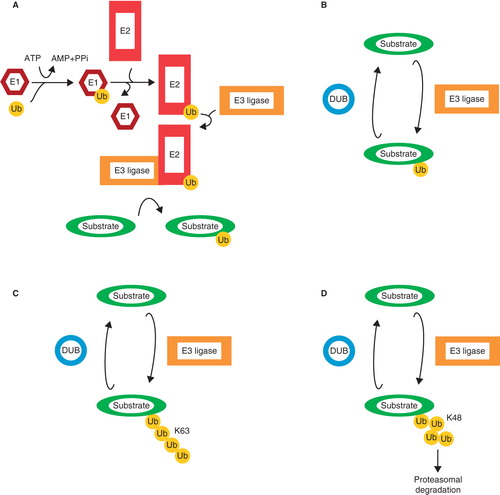
Figure 3. Positive regulation of TGFβ signaling by Arkadia. TGFβ signaling is inhibited by I-Smads, in co-operation with various E3 ligases, targeting several components among which the receptor complexes. SnoN and c-Ski inhibit TGFβ signaling at a later step by acting as transcriptional co-repressors. Arkadia targets I-Smad (Smad7), SnoN, and c-Ski for degradation, thereby positively regulating signaling.
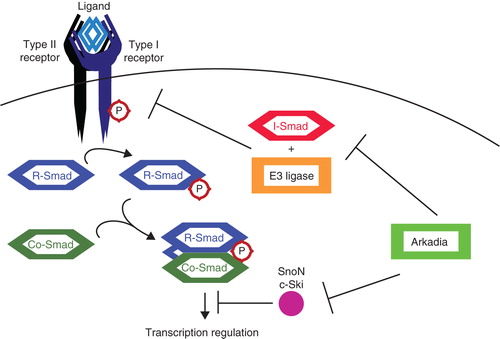
Figure 4. Mono-ubiquitination of Smad4. Active complexes of Co-Smad, Smad4, and phosphorylated R-Smads recruit histone acetyltransferases (HATs) to chromatin. The acetylation of histones recruits Ectodermin/TIF1γ (Ecto), which then disrupts Smad complexes and mono-ubiquitinates the Co-Smad. Released R-Smads are most likely dephosphorylated and exported to the cytoplasm. Ubiquitinated Smad4 is exported to the cytoplasm, where the deubiquitinating enzyme (DUB) FAM/USP9x removes the ubiquitin moiety. Smad4 is now ready once again to form complexes with R-Smads.
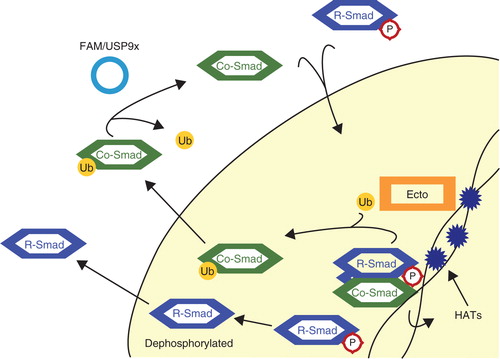
Figure 5. TRAF6 activates TAK1. Tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6) binds activated TGFβ receptor complexes and is activated via K63 autoubiquitination. TRAF6 subsequently ubiquitinates and activates TGFβ-associated kinase (TAK1), which is responsible for activating non-Smad pathways such as the p38 MAP kinase pathway.
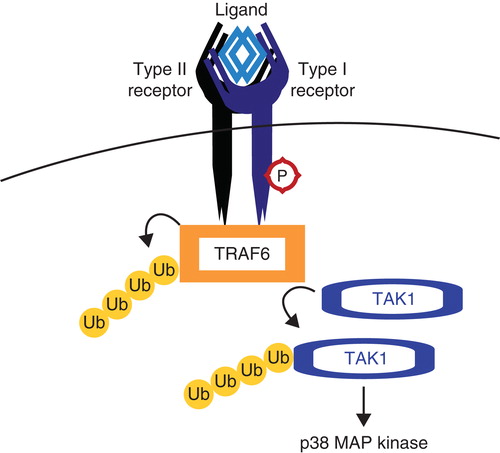
Table I. An overview of ubiquitin modifications involved in TGFβ signaling.