Figures & data
Figure 1. Decrease in phosphate in phosphoramidate phosphorylated histone H1 (▪) and 30 kDa polylysine (⧫) during incubation with PHPT1. The concentration was 1 mg/mL of phosphohistone and 2 mg/mL of phosphopolylysine. An amount of 5 pmol PHPT1 was added per 51 µL incubation volume. The reaction was performed at pH 7.5 and 30°C during indicated times and was interrupted by centrifugation of 50 µL of the reaction mixture through a spin column containing 200 µL of DEAE-Sepharose equilibrated in 25 mM Tris/HCl pH 8.5. The protein-bound, acid-labile phosphate in the final eluate was analyzed as described under Methods. Each time point was analyzed in duplicate.
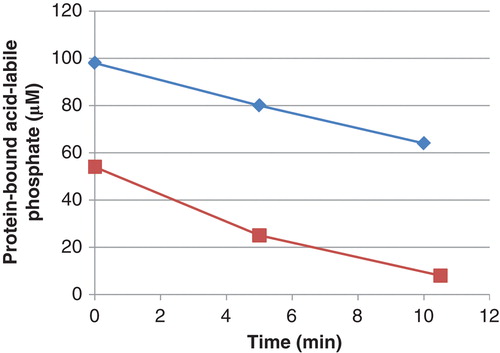
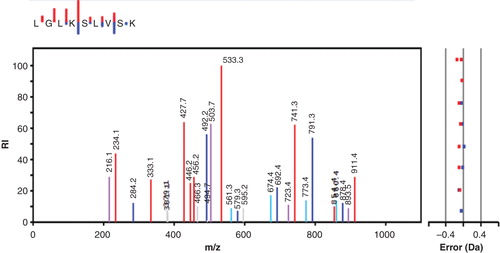
Figure 2. Amino acid sequence and phosphorylated sites of bovine histone H1.2 as determined by mass spectrometry. Lysine residues are marked red, and those identified as targets for the phosphorylation by phosphoramidate (i.e. seven residues) are highlighted. Out of the 305 peptides that were used to identify the protein, 14 peptides were reported to contain phospholysine.

Figure 3. MS/MS spectrum showing the fragmentation pattern of one of the peptides obtained after trypsin treatment of histone H1 from SignalChem. The y-ion series is shown in red, and the b-ions series is in blue. Also shown is y- and b-ions fitting with the loss of 18 Da in violet and turquoise, respectively. Other observed but unspecified mached ions are grey. The small inset at the top shows only the y- and b-ions, with the length of the staples representing the intensity of the ions.