Figures & data
TABLE 1. Characterization of subjects involved in the study
FIGURE 1. Melatonin profiles and their correlation with chronotype. (A) Grouped 24-h melatonin profiles for subjects with early (full circle) and late (full square) chronotypes. The data were expressed as the mean ± SEM (each chronotype n = 6) and fitted with a cosine curve to determine the acrophase of the rhythms. The acrophase of the early chronotypes (full line) occurred earlier than those of the late chronotypes (dashed line). (B) Relationship between acrophases of the 12 individual melatonin profiles (means ± SD) and their corresponding individual mid-sleep phases corrected for the sleep debt accumulated during workdays (MSFSC). The solid line shows the best-fit linear regression, the dashed lines represent 95% confidence intervals to the regression line, and R2 represents the goodness of fit. For details see Materials and Methods.
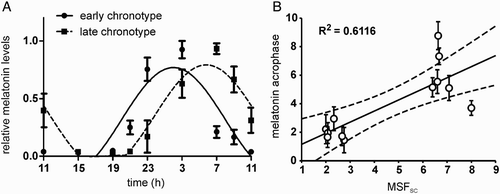
TABLE 2. Results of cosinor analysis of individual daily profiles of melatonin and clock gene expression levels
FIGURE 3. Clock gene expression profiles and their correlation with chronotype. Left panel: Grouped 24-h Per1 (upper), Per2 (middle), and Rev-erbα (lower) expression profiles for subjects with early (full circle) and late (full square) chronotypes. The data were expressed as the mean ± SEM (each chronotype n = 6) and fitted with a cosine curve to determine the acrophase of the rhythms. The acrophase of the early chronotypes (full line) occurred earlier than those of the late chronotypes (dashed line). Right panel: Relationship between the acrophases of the individual Per1 (upper), Per2 (middle), and Rev-erbα (lower) expression profiles (means ± SD) and the individual's mid-sleep phase corrected for the sleep debt accumulated during workdays (MSFSC). Only profiles with significant cosine fits (see Table 2) were considered for correlation (Per1: n = 11; Per2: n = 10; Rev-erbα: n = 9). The solid line shows the best-fit linear regression, the dashed lines represent 95% confidence intervals to the regression line, and R2 represents the goodness of fit. For details see Materials and Methods.
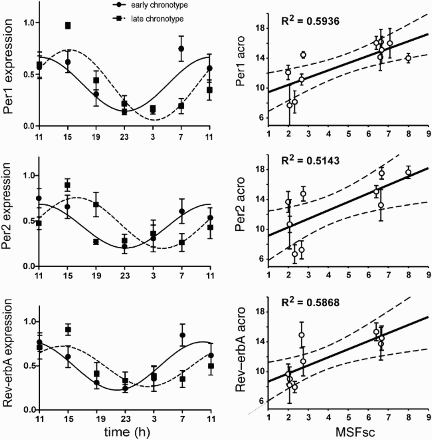
FIGURE 4. The relationship between the acrophases of the individual melatonin profiles and individual profiles of Per1 (upper), Per2 (middle), and Rev-erbα (lower) expression (means ± SD). Only profiles with significant cosine fits (see Table 2) were considered for correlation (Per1: n = 11; Per2: n = 10; Rev-erbα: n = 9). The solid line shows the best-fit linear regression, the dashed lines represent 95% confidence intervals to the regression line, and R2 represents the goodness of fit. For details see Materials and Methods.
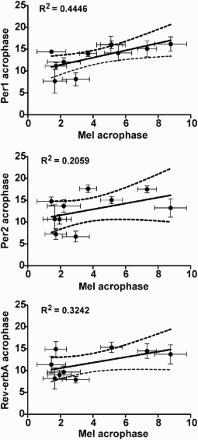
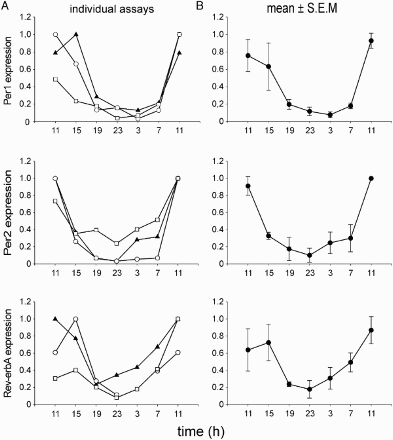
FIGURE 2. Evaluation of the method used for determining the phase of the clock gene expression profiles. Samples of buccal scrubs were collected throughout the 24 h in the same subject in three independent sessions. (A) Individual profiles of Per1 (upper), Per2 (middle), and Rev-erbα (lower) resulting from three independent assays. (B) Average profiles (means ± SEM.) of Per1 (upper), Per2 (middle), and Rev-erbα (lower) of the three measurements. For details see Materials and Methods.