Figures & data
Figure 1. Experimental design. A: Serum samples handling was optimized by removing salts and other potentially interfering substances. B: DIGE analyses revealed proteins differentially expressed in a significant manner between serum specimens from HCM and control patients. C: Preparative new gels were visualized using Coomassie staining for picking up selected spots. D: Proteins of interest were identified using liquid chromatography coupled with MS/MS. E: Nephelometry served to validate the HCM diagnostic properties of identified proteins on independent series of serum samples.
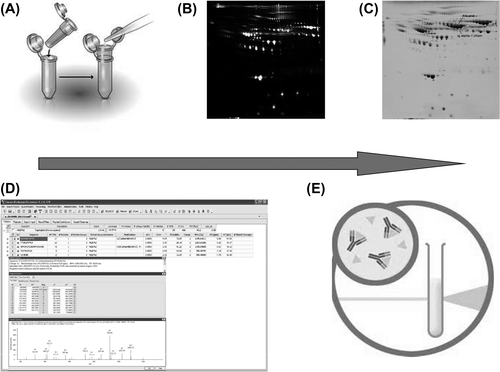
Table I. Information of patients and controls providing serum samples included for 2D-DIGE analysis.
Table II. Demographic and clinical characteristics of patients included in the 2D-DIGE analysis.
Figure 2. Representative 2D-DIGE staining of proteomic profiling from serum samples. A: Protein extract from HCM patients; and B: proteome extract from sex- and age-matched controls. Spots marked with a number indicate identified proteins whose abundance changes between HCM and control specimens reached statistical significance.
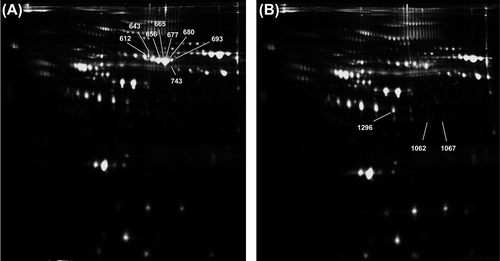
Table III. Demographic and clinical characteristics of patients included in the nephelometric immunoassay.
Figure 3. Three-dimensional view of differentially expressed proteins in serum samples from HCM patients compared with controls. Changes in protein expression from HCM and controls are shown by each line. The average ratio of expression for each selected protein grouping in HCM and control specimens as obtained by computational analysis with DeCyder Software is represented by lines marked with a cross. Statistical analysis allowed detection of significant abundance changes (95th confidence level) based on the variance of the mean change within the cohort: A: haptoglobin; B: hemopexin; C: Ig alpha-1 chain C region isoform; D: albumin.
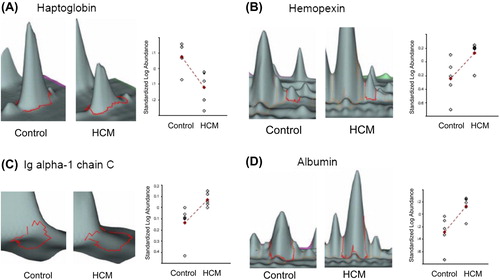
Table IV. Significant differentially expressed proteins identified by reverse-phase liquid chromatography coupled with MS/MS after 2D-DIGE analysis of HCM patients versus control serum samples.
Table V. Correlation of haptoglobin and hemopexin serum levels with HCM.