Figures & data
Figure 1. Physiological processes regulating the sleep-wake cycle and indicator of sleep propensity. (A) The sleep-wake cycle is influenced by two main regulatory processes. The circadian process is mainly orchestrated by the master pacemaker located in the suprachiasmatic nuclei (SCN) of the hypothalamus and drives a circadian (about 24 h) rhythm in sleep propensity. The homeostatic process is represented by the build-up of sleep need during wakefulness and its decay in the course of sleep. (B) It is commonly accepted that slow-wave activity (SWA; 0.75–4.5 Hz EEG activity) during non-rapid eye movement sleep (NREMS) is a key marker of the homeostatic process because it increases as a function of wakefulness duration and dissipates during sleep, reflecting the decay in homeostatic sleep need. During sleep deprivation, high sleep pressure leads to increased SWA that returns to baseline level during the recovery sleep. (EEG = electroencephalographic; REMS = rapid-eye-movement sleep).
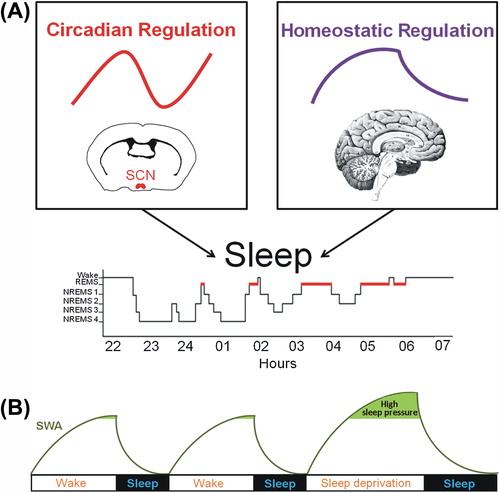
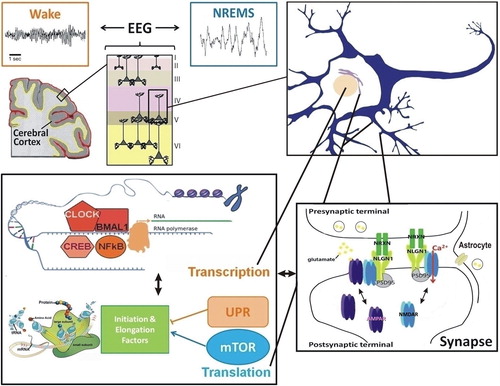
Figure 2. Interconnections between molecular elements linked to sleep homeostasis. The EEG reflects the synchronization of neuronal firing in the cerebral cortex to which pyramidal cells, among others, contribute. Changes in neuronal firing synchrony across the sleep-wake cycle, which occur together with changes in neurotransmitters (e.g. glutamate, acetylcholine, dopamine, serotonin), depend on modifications in individual neurons leading to modifications in properties of neuronal communication (e.g. synaptic function). At the transcriptional level, numerous transcription factors seem to respond to neuronal activity associated with prolonged wakefulness and to impact on EEG activity during wakefulness and sleep (e.g. NFκB, CLOCK/BMAL1, CREB). Protein regulation also seems a relevant cellular function linked to sleep, particularly in the context of memory consolidation. Indeed, the mTOR pathway, involved in protein synthesis, and the unfolded protein response (UPR), linked to protein folding and degradation, are usually associated with the sleep and wake states, respectively. Importantly, transcriptional and translational machineries are interacting such that many of these pathways were shown to influence each other directly. In addition, these interactions may feed back on synaptic properties to determine the function of the synapses, notably glutamatergic synapses, as a function of sleep need. This likely involves neurons and non-neuronal cells like astrocytes, and leads to the modulation of numerous synaptic components (e.g. NMDA and AMPA receptors, Neuroligins). Such cross-talk between these molecular and cellular levels of regulation accounts for the changes in EEG synchrony as a function of the sleep/wake history. (EEG = electroencephalogram; mTOR = mammalian target of rapamycin; NLGN1 = Neuroligin-1; NREMS = non-rapid-eye-movement sleep; NRXN = Neurexin; PSD95 = postsynaptic density protein 95; UPR = unfolded protein response).