Figures & data
Figure 1. Summary of chimeras used to clarify the role of the N-terminus of SERCA1 in ER retrieval. All of the chimeras were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences.
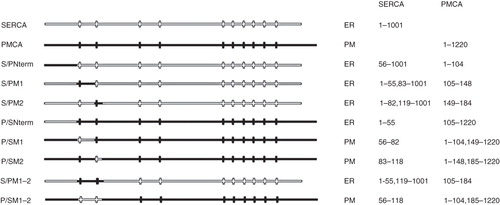
Figure 2. Expression of EGFP-tagged SERCA1 and PMCA3 by COS-7 cells. COS-7 cells were transfected with DNA encoding for SERCA1-EGFP (a–c) and PMCA3-EGFP (d–f) in the expression vector pcDNA3.1(+). After two days the cells were treated with brefeldin A, fixed in methanol, permeabilized with triton X100, before treating with antibodies directed against TGN46, followed by a Texas Red conjugated secondary antibody. Laser scanning confocal images of EGFP are shown in panels a and d, Texas Red in b and e and overlaid images in c and f. Scale bars are indicated.
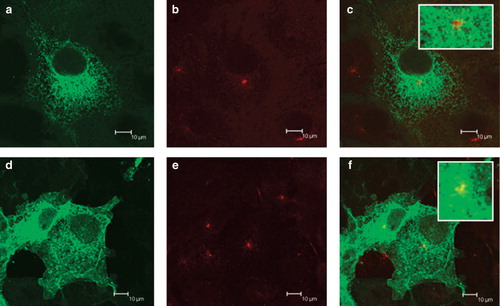
Figure 3. Location of chimeras used to examine the role of the N-terminus of SERCA1 in ER retrieval. COS-7 cells were transfected with the chimeric constructs all tagged at their C-terminus EGFP: P/SM1 (panels a–c); P/SM2 (panels d–f); S/PM1-2 (panels g–i); P/SM1–2 (panels j–l). After two days the cells were treated with brefeldin A, fixed in cold methanol, and permeabilized with triton X100 before incubating with antibodies against TGN46, visualized with Texas Red conjugated to a secondary antibody. Confocal images of EGFP fluorescence are shown in panels a, d, g, j, m, p, s and v. Texas Red images are shown in panels b, e, h, k, n, q, t and w. The merged images are shown in panels c, f, i, l, o, r, u and x.
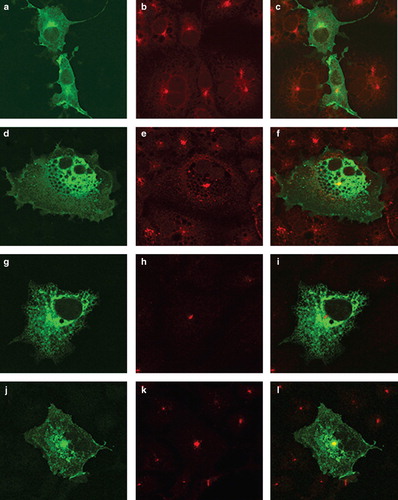
Figure 4. Summary of chimeras used to examine the role of SERCA1 C-terminus in ER retrieval. All of the chimeras were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences.
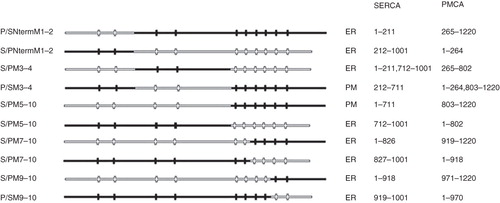
Figure 5. Location of chimeras used to examine the role of the C-terminus of SERCA1 in ER retrieval. (a) COS-7 cells were transfected with the chimeric constructs all tagged at their C-terminus with EGFP: P/SNtermM1–2 (panels a–c); S/PNtermM1–2 (panels d–f); S/PM3–4 (panels g–i); P/SM3–4 (panels j–l); S/PM5–10 (panels m–o); P/SM5–10 (panels p–r). After two days the cells were treated with brefeldin A, fixed in cold methanol, and permeabilized with triton X100 before incubating with antibodies against TGN46, visualized with Texas Red conjugated to a secondary antibody. Confocal images of EGFP fluorescence are shown in panels a, d, g, j, m and p. Texas Red images are shown in panels b, e, h, k, n and q. The merged images are shown in panels c, f, i, l, o, and r. The images from panels 5 c, f, i, l, o and r were analyzed using ImageJ software from NIH (http://rsbweb.nih.gov/ij/). The profiles of fluorescence intensity across the TGN for EGFP and Texas Red fluorescence are plotted in green and red respectively.
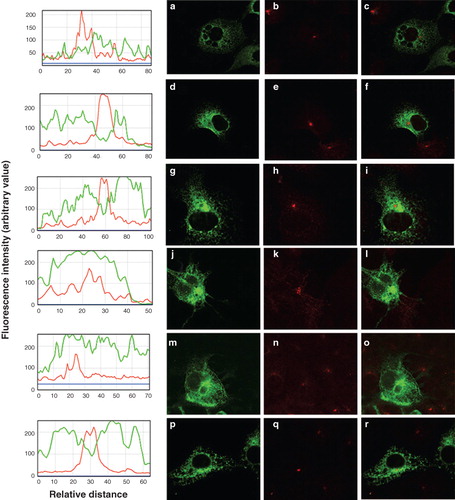
Figure 6. Summary of other constructs used to identify the location of the C-terminal ER retrieval sequence of SERCA1. All of the constructs were tagged with EGFP at their C-termini. The designated names are provided in the left column and their proposed locations, from analysis of co-localization studies, are indicated as well the precise sequences making up the chimeras. The filled horizontal lines represent PMCA3 sequence and the unfilled lines SERCA1 sequence. The vertical lines indicate the approximate locations of the transmembrane sequences. The exceptions are construct CD8, which contains the human CD8 sequence and CD8-SERCA M10 and CD8-PMCA M10 in which the transmembranous domain of CD8 is replaced by M10 of SERCA1 and PMCA3, respectively. The dashed lines in constructs SM1–2M9–10 and SM1–2/PM9–10 represent flexible linkers between the SERCA1 and PMCA3 sequences.
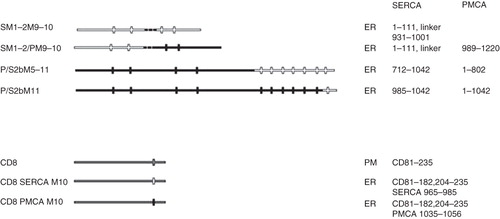
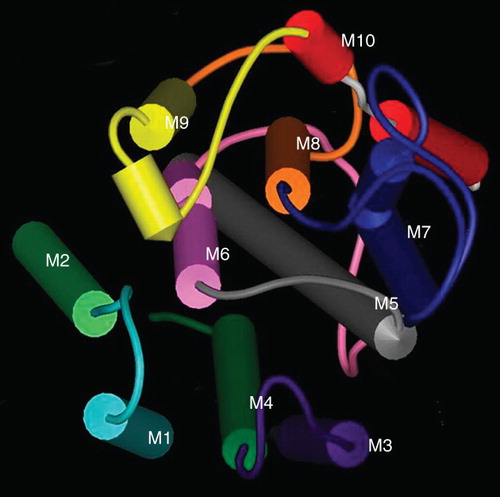
Figure 7. The organization of the transmembrane helices of SERCA1. A space fill model of the transmembrane domains of SERCA1 produced using the coordinates deposited in the Protein Data Base (ID 1SU4). The helices have been colour-coded M1 light blue, M2 light green, M3 purple, M4 dark green, M5 grey, M6 pink, M7 blue, M8 orange, M9 yellow, M10 red. The view is from the ER lumen.