Figures & data
Figure 1. Topology model of class ST[3] transporters. The membrane topology of GltS of E. coli is shown which is representative for the core structure of class ST[3] transporters. Cylinders represent transmembrane segments. The homologous N and C domains that have opposite orientation in the membrane were indicated in dashed boxes. VB and XA represent the reentrant loops in the N- and C-domain, respectively. Black dots point at the positions of mutation P351C and N356C introduced by site directed mutagenesis. The open square points at position 206, the insertion site of the BAD domain in the hybrid version of the GltS protein.
![Figure 1. Topology model of class ST[3] transporters. The membrane topology of GltS of E. coli is shown which is representative for the core structure of class ST[3] transporters. Cylinders represent transmembrane segments. The homologous N and C domains that have opposite orientation in the membrane were indicated in dashed boxes. VB and XA represent the reentrant loops in the N- and C-domain, respectively. Black dots point at the positions of mutation P351C and N356C introduced by site directed mutagenesis. The open square points at position 206, the insertion site of the BAD domain in the hybrid version of the GltS protein.](/cms/asset/b7faf126-2dc0-4ddc-a48c-ed872c3491ec/imbc_a_624989_f0001_b.jpg)
Figure 2. Effect of insertion of the BAD domain in the central loop on L-glutamate uptake activity of GltS. A,B. RSO vesicles expressing GltS (▪), GltS BAD206 (□) or control vesicles (▴) were assayed for L-glutamate (A) and proline (B) uptake activity. (C) SDS-PAGE of GltS (lane 1) and GltS BAD206 (lane 2) partially purified from the same batch of vesicles used in the uptake experiments shown in panels A and B. Molecular masses of marker proteins (lane M), masses were indicated in kDa. (D) Relation between uptake activity and expression of GltS (▪) and GltS BAD206 (□) in RSO membranes. L-glutamate uptake was measured in RSO membranes isolated from cells induced with 0, 0.001 and 0.1% (w/v) of arabinose. Expression levels were determined densitometrically after partial purification of the transporter proteins from the same RSO membranes followed by SDS-PAGE. The insert shows the Coomasie Brilliant Blue (CBB) stained gel.
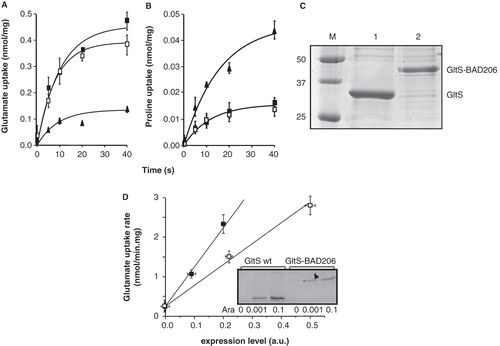
Figure 3. Kinetic parameters of GltS in RSO membranes. The initial rate of uptake of L-glutamate at a concentration of 2.9 μM in RSO membranes expressing GltS was measured at the indicated Na+ concentrations. The K0.5 for Na+ was 5.8 ± 0.5 mM. Inset, L-glutamate dependent kinetics of the same membranes at a concentration of 10 mM of Na+. The affinity for glutamate was KM = 13.5 ± 3.5 μM.
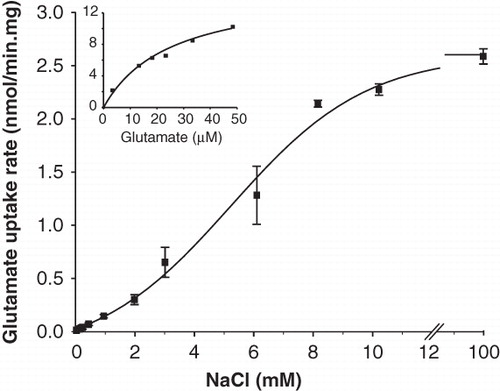
Table I. Kinetic parameters of GltS and GltS-BAD206 for pmf driven L-glutamate uptake in RSO membranes.
Figure 4. Sensitivity of P351C and N356C mutants of GltS to inactivation by NEM and AmdiS. RSO membranes containing mutants GltS-N356C and BAD206-N356C (A) and GltS-P351C and BAD206-P351C (B,C) were treated in 50 mM KPi pH 7 buffer for 10 min with NEM (A,B) and AmdiS (C) at the indicated concentrations. Following quenching of the remaining thiol reagent by access DTT, washing of the membranes, and resuspension in 50 mM KPi pH 6 buffer, residual L-glutamate uptake activity was measured relative to membranes containing the same mutant that was treated in exactly the same way, except that the thiol reagents were omitted during the treatment. Open bars, GltS-N356C and GltS-P351C. Shaded bars, BAD206-N356C and BAD206-P351C. Back ground activities amounted to 30, 25, 20 and 50% for the membranes containing mutants GltS-N356C, BAD206-N356C, GltS-P351C and BAD206-P351C, respectively.
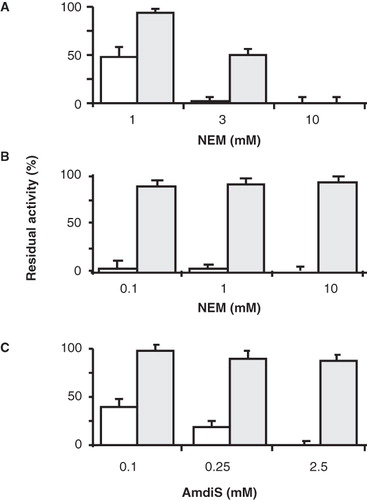
Figure 5. Protection of GltS-P351C (A) and GltS-N356C (B) mutants by L-glutamate. RSO vesicles containing GltS-P351C (A) or GltS-N356C (B) were treated in 50 mM KPi pH 7 buffer without (▪) and with 0.25 mM AmdiS (A) or 1 mM NEM (B) for 10 min in the presence (▴) or absence (•) of 1 mM L-glutamate. The treatment was stopped by the addition of 1 mM DTT. L-glutamate uptake was measured after extensive washing of the membranes to remove unlabeled L-glutamate and resuspension in 50 mM KPi pH 6 buffer.
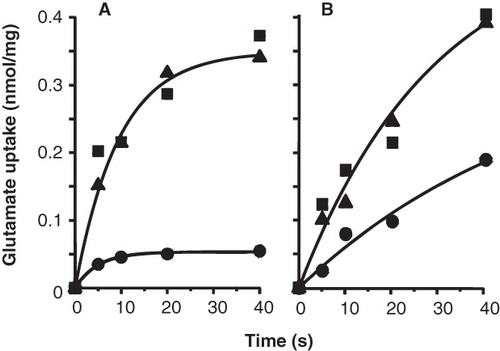
Figure 6. Effect of the co-ion Na+ on the sensitvity of the P351C and N356C mutants to thiol reagents. RSO membranes containing GltS-P351C and BAD206-P351C (A) and GltS-N356C and BAD206-N356C mutants were treated with AmdiS or NEM in 50 mM KPi pH 7 buffer containing 0, 10 and 100 mM NaCl as indicated. GltS-P351C (open bars) and the BAD206-P351C (shaded bars) were treated with 0.25 mM AmdiS for 10 min. GltS-N356C (open bars) was treated with 1 mM NEM for 10 min and the BAD206-N356C (shaded bars) with 3 mM NEM for 20 min. Following the treatment, the membranes were washed and resuspended in 50 mM KPi pH 6 buffer containing 10 mM Na+. Bars indicate the L-glutamate uptake activity relative to a sample that was not treated with the thiol reagents.
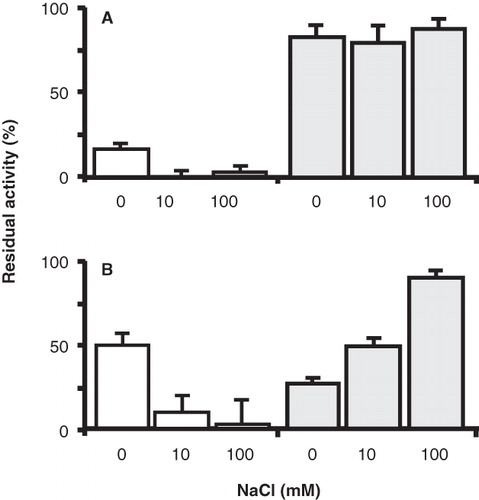
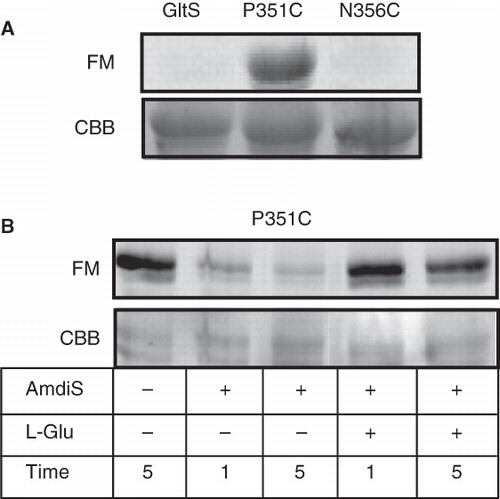
Figure 7. Labeling of GltS and the GltS-P351C and GltS-N356C mutants by fluorescein maleimide in whole cells. A. Whole cells expressing GltS, GltS-P351C, and GltS-N356C were treated for 1 min with 0.1 mM fluorescein maleimide. B. Cells expressing GltS-P351C were treated for the indicated times with 0.25 mM AmdiS and in the presence or absence of 1 mM L-glutamate as indicated. Following quenching and removal of the excess thiol reagent, the cells were treated with fluorescein maleimide as described under A. Following the treatment, the transporter proteins were isolated from the cells and analyzed by SDS-PAGE. The lower panels (CBB) show the Coomasie Brilliant Blue stained protein bands, the top panels the fluorescence image of the same parts of the gels (FM).