Figures & data
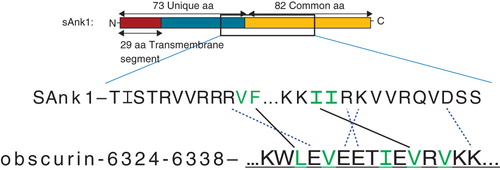
Figure 1. Images of sAnk-1 alone and the sAnk-157–122:Obsc6322–6339 complex. (A) Top and (B) side views of the hydrophobic hotspot within the binding region of sAnk1, from the 30 ns time frame of the sAnk-1 monomer MD simulations. The backbone is shown in red cartoon representation while the four surface-exposed hydrophobic residues: Val-70 (green), Phe-71 (yellow), Ile-102 (blue), and Ile-103 (red) are shown as surface representations, (C) Front and (D) rear views of the sAnk-157–122:Obsc6322–6339 complex. sAnk-1 is shown in a red cartoon representation along with the four residues shown in panels A and B; obscurin is shown in a blue cartoon representation with L6326 (green), V6328 (yellow), I6332 (blue), V6334 (red) and V6336 (cyan) shown as a surface representation. Full solvent accessible surfaces representations of both proteins are included as white transparent surfaces. Structures for panels C and D were from the 7.7 ns time frame of the 30 ns MD simulation of wild type sAnk1 in complex with Obsc6322–6339, which has an average VDW interaction energy of 1.62 kcal/mol between I102 of sAnk1 and V6334 of obscurin.
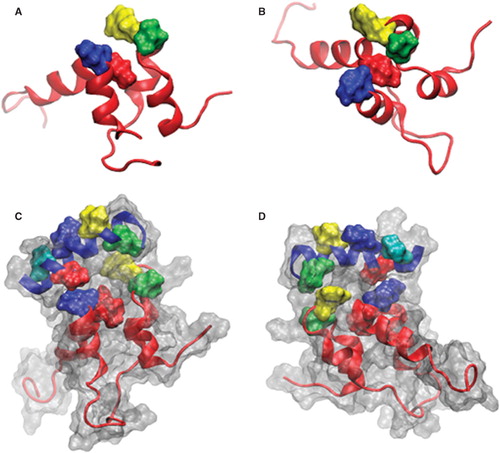
Figure 2. Alanine mutations within the hydrophobic hotspot of sAnk1-MBP reduce binding to GST-Obsc6316–6345. (A) The nitrocellulose membrane following transfer from the SDS-PAGE gel was stained with Ponceau Red confirmed equal loading and transfer of GST and GST-Obsc6316–6345. (B) Site-directed mutagenesis was used to create the four alanine-substituted MBP fusion protein mutants of sAnk1. Coomassie Blue stain shows the affinity-purified proteins, after analysis at equal load by SDS-PAGE. (C) For the blot overlay, a ladder of protein standards was run in lane 1 and the 25 kDa marker is shown. The remaining even lanes were loaded with GST and odd lanes with GST-Obsc6316–6345, at equal protein loads. The resulting gel was transferred to nitrocellulose. Each pair of lanes was then overlaid individually with each of the five sAnk1-MBP fusion proteins from B and probed with anti-MBP, followed by a fluorescent secondary antibody. The results show that each of the four alanine mutants of sAnk1 reduces binding to GST-Obsc6316–6345. Binding to each construct is specific, as none of the sAnk1 constructs bind to GST alone.
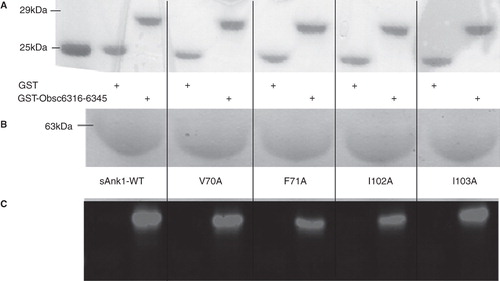
Figure 3. Representative SPR traces of GST-Obsc6316–6345 binding to wild type and mutant sAnk1. Four serial dilutions of sAnk1 (A) and the mutants (B-E), at concentrations of 1 μM and lower, were prepared and flowed over chip surfaces bound with GST-Obsc6316–6345 for 3 min, before dissociation. 1 μM is shown in black as the top curve followed by 500 nM, 250 nM and the lightest shade of grey at the bottom is 125 nM. The overall fit of the binding curves was based on the 1:1 Langmuir model provided by Biaeval software.
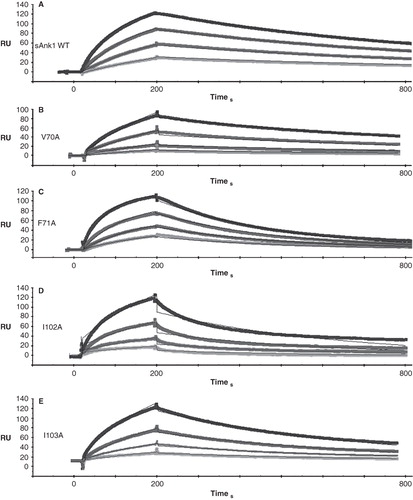
Figure 4. Quantitation of the kinetics for binding of hydrophobic mutants of sAnk1 to Obsc6316–6345. Surface plasmon resonance (SPR) was used to measure the binding kinetics of the high affinity binding site Obsc6316–6345 to wild type sAnk1 and mutants in which the hydrophobic residues were converted to alanines. (A) The on rate, kon, for all mutants was significantly decreased compared to wild type. (B) The dissociation rate, koff, for all of the mutants except V70A was significantly increased, compared to wild type. (C) The calculated KD's, based on the results in A and B, show that all mutants result in a decreased overall binding affinity compared to wild-type sAnk1. The F35A mutant protein was used as a control to show the alanine mutations are specific to the hydrophobic hotspot. n = 12 for WT, n = 9 for V70A, n = 5 for F71A, I102A, and I103A, and n = 4 for F35A. * = p < 0.05.
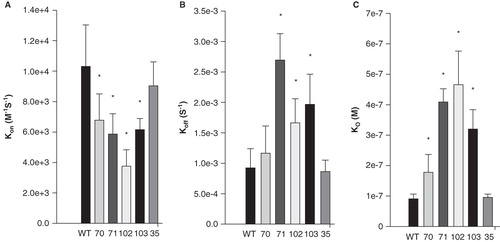
Figure 5. sAnk1-MBP mutations of two hydrophobic residues in hotspot reduce binding to GST-Obsc6231–6260. Methods were the same as in , except site-directed mutants of Obsc6231–6260 were assayed. As in , even lanes were loaded with GST and odd lanes with GST-Obsc6231–6260, at equal protein loads, except lane 1, which shows the standard proteins. Qualitatively, a reduction in binding is observed in lanes 7 and 11 corresponding to decreased binding affinity of F71A and I103A for Obsc6231–6260.
Figure 6. Representative SPR traces of GST-Obsc6231–6260 binding to wild type and mutant sAnk1. Experiments were performed as stated in , with GST-Obsc6231–6260 attached to the chip surface and sAnk1 analytes applied at serial dilutions from 1 μM (top curve in black) to 125 nM (bottom curve light grey).
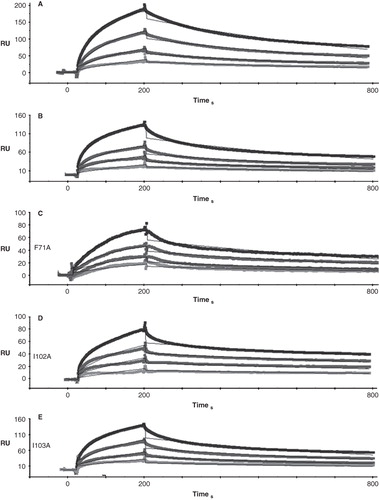
Figure 7. Quantitative evaluation of the binding kinetics for sAnk1 constructs binding to Obsc6231–6260. SPR was used to measure the binding kinetics of this region of obscurin to the wild type sAnk1 and its hydrophobic alanine mutants, as in , but the Obsc6231–6260 sequence was assayed. (A) The association rate, kon, for the F71A and I103A mutants of sAnk1, but not for the V70A or I102A mutants, was significantly decreased compared to wild type. (B) The dissociation rate, koff, for the mutants was not significantly different for binding to Obsc6231–6260 compared to wild type (p = 0.44 for V70A, 0.69 for F71A, 0.18 for I102A and 0.31 for I103A). (C) The calculated overall binding constants, KD, based on the results in A and B, show only the F71A and I103A mutants of sAnk1 with decreased binding affinities, compared to wild type. n = 4 for all panels, * = p < 0.05.
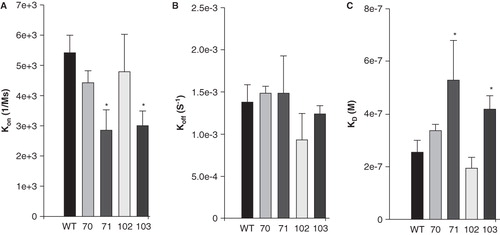
Figure 8. Mutations of three hydrophobic residues within GST-Obsc6316–6345 reduce binding to sAnk1-MBP. (A) The primary sequence of this region of obscurin contains five hydrophobic residues within the binding site (Leu-6326, Val-6328, Ile-6332, Val-6334 and Val-6336), all highlighted. (B) Blot overlay was used to assay the binding of each of the alanine mutants to wild type sAnk1-MBP. Lanes: (1) Standard; (2) GST; (3) GST-Obsc6316–6345; (4) GST-Obsc6316–6345-L6326A; (5) GST-Obsc6316–6345-V6328A; (6) GST-Obsc6316–6345-I6332A; (7) GST-Obsc6316–6345-V6334A; and (8) GST-Obsc6316–6345-V6336A.
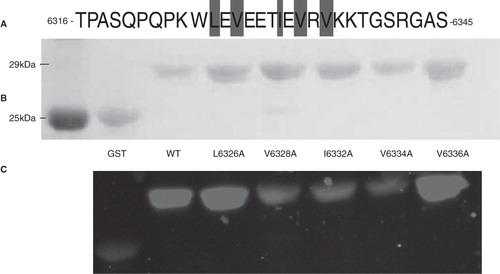
Figure 9. Multiple but not single alanine mutants of GST-Obsc6231–6260 reduce binding to sAnk1-MBP. (A) This region of obscurin has six hydrophobic residues located toward the N-terminus (Val-6233, Ile-6234, Ile-6235, Ile-6239, Val-6242 and Val-6243), all highlighted. (B) Blot overlays were used to assay the ability of each alanine mutant within Obsc6231–6260 to bind to wild type sAnk1-MBP. In addition, a triple mutant of the three N-terminal hydrophobic residues, as well as a double mutant of the last two neighboring hydrophobic residues, were tested. Lanes: (1) Standards; (2) GST; (3) GST-Obsc6231–6260; (4) GST-Obsc6231–6260-V6233A; (5) GST-Obsc6231–6260-I6234A; (6) GST-Obsc6231–6260-I6235A; (7) GST-Obsc6231–6260-I6239A; (8) GST-Obsc6231–6260-V6242A; (9) GST-Obsc6231–6260-V6243A; (10) GST-Obsc6231–6260-V6233A/I6234A/I6235A; and (11) GST-Obsc6231–6260-V6242A/V6243A.

Table I. Per-residue solvent accessibility (SA) of wild type and mutant sAnk1 as a monomer and in complex with obscurin. ΔSA = SA(Complex) − SA(Monomer).
Table II. VDW interaction energies between hydrophobic residues in sAnk1 and obscurin tested experimentally.
Figure 10. Hydrophobic character at position 102 of sAnk1 is necessary for high affinity binding to Obsc6316–6345. SPR measured the binding kinetics for sAnk1 constructs with different amino acid residues at position 102. (A) The on rate, kon, for wild type sAnk1, sAnk1-I102F and -I102L show no differences in association profiles for Obsc6316–6345. (B) The dissociation rate, koff, for the mutants are not significantly increased compared to wild type. Rather, the dissociation rate of sAnk1-I102F for binding to Obsc6316–6345 is significantly decreased. (C) The calculated overall binding constant, KD, based on the results in A and B. Both I102F and I102L retain high affinity binding for Obsc6316–6345 while sAnk1-I102F forms a stronger complex than wild type. sAnk1-I102D did not produce curves that could be analyzed, due to poor association and dissociation. n = 5 for WT sAnk1, sAnk1-I102F and sAnk1-I102L, n = 3 for sAnk1-I102D, * = p < 0.05. (D) Blot overlay assays of GST (lanes 2, 4, 6, 8) and GST-Obsc6316–6345 (lanes 3, 5, 7, 9). Lane 1 shows standard proteins. Lanes 2–3 were probed with wild-type sAnk1, lanes 4–5 with sAnk1-I102F, lanes 6–7 with I102L, and lanes 8–9 with sAnk1-I102D.
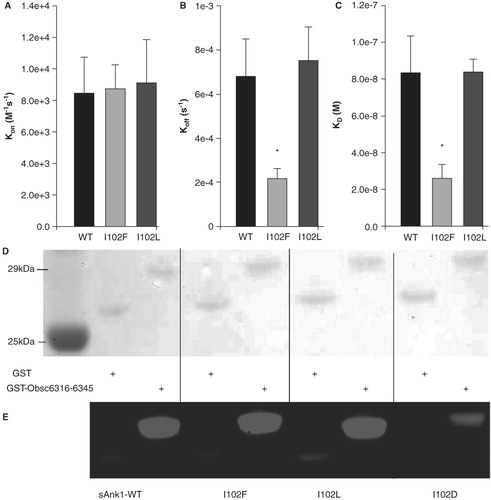
Figure 11. Residues involved in binding between the high affinity binding site of obscurin and sAnk1. Our lab has previously identified charged pairs of amino acids involved in binding between these proteins. The binding residues are matched to their partner with dotted lines. The hydrophobic residues necessary to preserve high affinity binding are highlighted in green. In addition, the favorable interactions between I102/V6334 and F71/L6326 as observed during MD simulations and I102/V6334 as confirmed biochemically, are shown with solid lines to highlight how high affinity binding is achieved.