Figures & data
Figure 1. Exponential growth of the number of unique membrane protein X-ray structures available since 1985. Produced using data from: http://blanco.biomol.uci.edu/mpstruc/listAll/list.
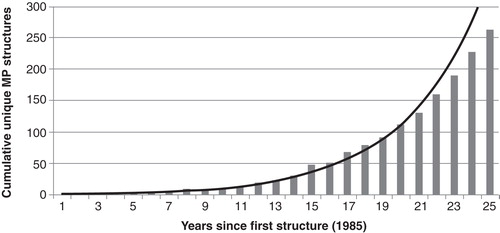
Figure 2. LeuT structure. (a) Overall structure of the LeuT protein with the S1-binding site containing L-leucine (green) and two sodium ions (magenta) and the S2-binding site containing the tricyclic antidepressant imipramine (cyan). (b) Close-up of the S1-binding site; the leucine molecule sits in a very tight binding site and makes several hydrogen bonding interactions with Ala22, Gly26, Thr254 and Ser256. There is also a large hydrophobic interaction between the ligand and Tyr108, Phe259 and Ile359. This Figure is reproduced in colour in Molecular Membrane Biology online.
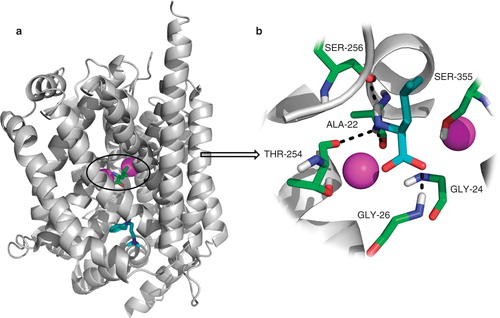
Figure 3. Schematic overview of the eHiTS docking strategy. In eHiTS, ligands are divided into rigid fragments and connecting flexible chains (Step 1). These fragments are docked individually into the binding site of the target protein (Step 2) and a fast graph-matching algorithm finds all matching solutions to reconstruct the original molecule (Step 3), which can then be optimized (Step 4), scored and ranked (Step 5). Reproduced with permission from Nature Reviews Microbiology (Simmons et al. Citation2010). This Figure is reproduced in colour in Molecular Membrane Biology online.
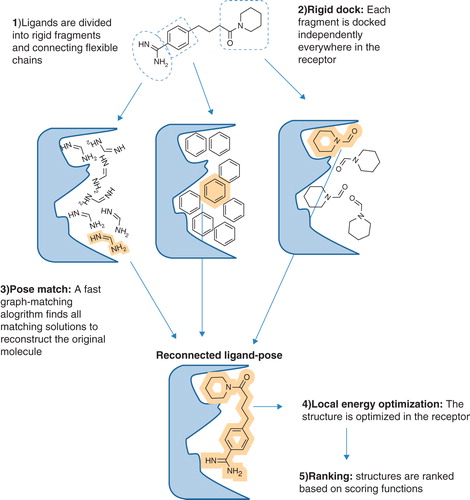
Figure 4. vHTS screening cascade. Starting from a commercially available compound library of 300,000 compounds, initial vHTS identified 1000 compounds which were evaluated further using consensus scoring and also a knowledge of compound solubility. A total of 11 compounds were purchased from Maybridge and evaluated for activity against the LeuT. This Figure is reproduced in colour in Molecular Membrane Biology online.
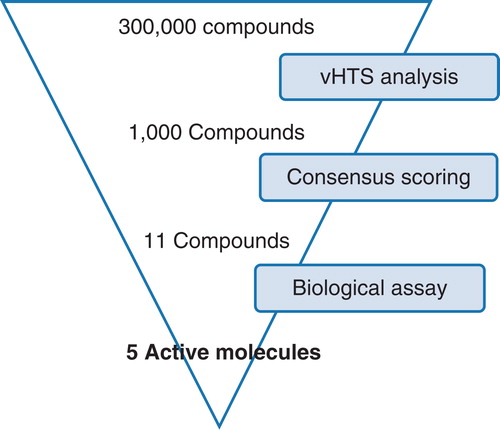
Figure 5. Dose-response inhibition of [3H]-leucine ([3H]-Leu) binding to LeuT by compounds 2, 4, 8 and 9. Normalized results are given as mean ± SEM on the basis of four independent experiments.
![Figure 5. Dose-response inhibition of [3H]-leucine ([3H]-Leu) binding to LeuT by compounds 2, 4, 8 and 9. Normalized results are given as mean ± SEM on the basis of four independent experiments.](/cms/asset/23a078be-2f98-407b-922c-3589b6b7665b/imbc_a_710341_f0005_b.jpg)
Table I. Identification of inhibitors of LeuT. % of total [3H]-leucine binding in the presence of 1 mM tested compounds and IC50 values for the inhibition of [3H]-leucine binding by compounds 2, 4, 8 and 9 or clomipramine (CMI) are given as mean ± SEM. Numbers in parentheses (N) indicate number of independent experiments.
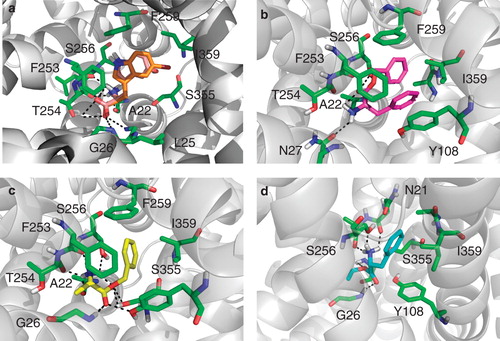
Figure 6. Binding pose of compound 2, 4, 8 and 9. (a) Compound 2 (orange) overlaid with its inactive variant compound 10 (salmon) docked into the crystal structure of the LeuT bound to L-tryptophan and sodium ions (PDB code 3F3A). The compounds makes H-bonding interactions with Leu25, Gly26 and Thr254 as well as filling a hydrophobic pocket created by Phe259 and Ile359, this pocket is violated by compound 10. (b) Compound 4 (magenta) docked into the crystal structure of the LeuT used for all vHTS (2Q72). The compound makes H-bonding interactions with Ala22, Asn27 and Ser256. (c) Compound 8 (yellow) docked into the crystal structure of the LeuT used for all vHTS (2Q72). The compound makes H-bonding interactions with many active site residues including Ala22, Tyr108 and Ser256. (d) Compound 9 (cyan) docked into the crystal structure of the LeuT used for all vHTS (2Q72). The compound makes H-bonding interactions with Asn21, Ala22, Gly24 and 26 and Ser256. This Figure is reproduced in colour in Molecular Membrane Biology online.