Figures & data
Table I. Characteristics of the dipeptide sequences selected.
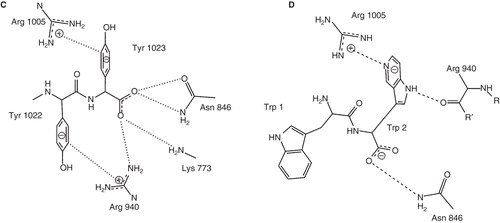
Figure 1. The Na,K-ATPase C-terminal structure and the functional cycle. (A) The structural context of the two C-terminal tyrosines (in spheres) of the Na,K-ATPase α-subunit (brown). The β-subunit (purple), the γ-subunit (green), the two occluded potassium ions (red) and the approximate position of the membrane (grey) are also indicated. The Figure was made with pymol (www.pymol.org) using PDB ID 2ZXE (a shark Na,K-ATPase). (B) The Post-Albers scheme for the Na,K-ATPase functional cycle. In E1P (top left), three sodium ions (green circles) are bound in the occluded pump. With a conformational change to the E2P state (top middle), a proton (black circle) approaches site IIIb from the cytoplasm, the extracellular gate opens, and the sodium ions are released. With a proton at the aspartate in site IIIb (Asp933 in Homo sapiens α1), two potassium ions (red squares) can bind, which promotes dephosporylation to the occluded E2 state (top right). Release of potassium and proton depends on opening of the C-terminal channel between TM5, TM7 and TM8 (bottom right). Binding of sodium and ATP (bottom middle) leads to E1 (bottom left). Figure adapted from Poulsen et al. (Citation2010) with the author's permission. (C) ChemDraw illustration of the interactions of the C-terminal tyrosines in 2ZXE. The program Capture “http://capture.caltech.edu/” suggests a single significant cation-π interaction between Arg940 and Tyr1022. Furthermore, Arg1005 is only 4.3 Å from Tyr1023, and mutational studies show decreased sodium affinity for Arg1005Gln (Poulsen et al. Citation2010), indicating that this may also be a relevant cation-π interaction, at least in part of the catalytic cycle. (D) ChemDraw illustration of the interactions suggested by Vina of the tryptophane-tryptophane dipeptide. In both cases, the most important interactions are between arginines and aromatic systems. This Figure is reproduced in colour in Molecular Membrane Biology online
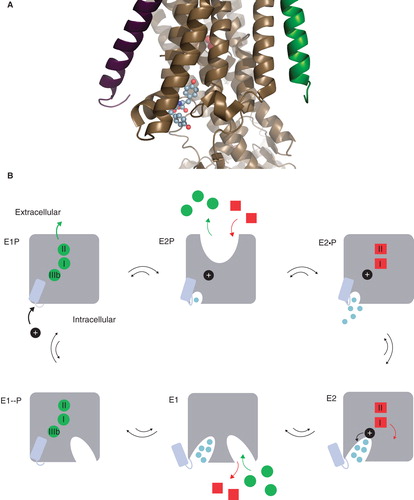
Figure 2. The voltage dependence of transient currents of C-terminally mutated Na,K-ATPases. The charge translocation (Q/Qmax) of C-terminal mutants was determined from the off pulses of a series of 20-mV voltage steps between −200 mV and 40 mV and Boltzmann fitted. Numbers in brackets indicate the V0,5 for extracellular Na+. A full list of V0,5 and slope for each of the tested constructs can be found in . This Figure is reproduced in colour in Molecular Membrane Biology online
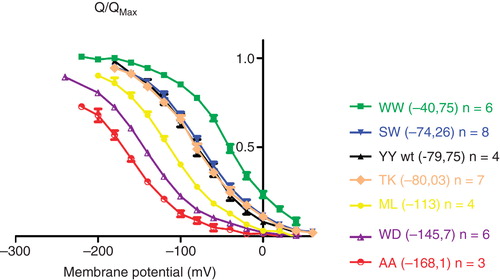
Figure 3. Comparisons of docking scores and sodium affinities. The scores for each of the four docking methods as well as the consensus score were plotted against the V0.5 values for the C-terminal mutants. The wild-type YY is green. For each of the plots, the Spearman correlation coefficient is indicated. There is a significant correlation for Vina, Glide EM and the consensus score (p < 0.05). The statistical analyses were done with GraphPad Prism. This Figure is reproduced in colour in Molecular Membrane Biology online
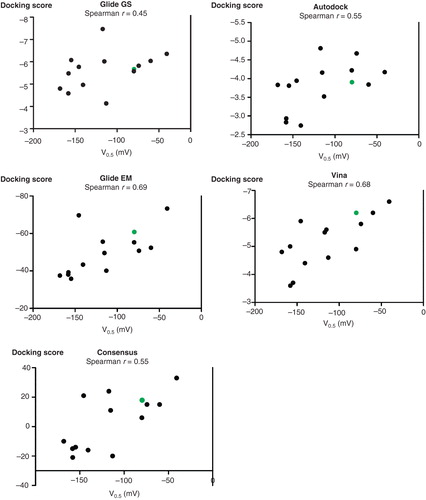
Figure 4. The voltage dependence of the relaxation rates of C-terminally mutated Na,K-ATPases. From single exponential fittings of the on pulses of a series of voltage steps between −180 mV and 40 mV, the relaxation rate constants of C-terminal mutants were determined. This Figure is reproduced in colour in Molecular Membrane Biology online
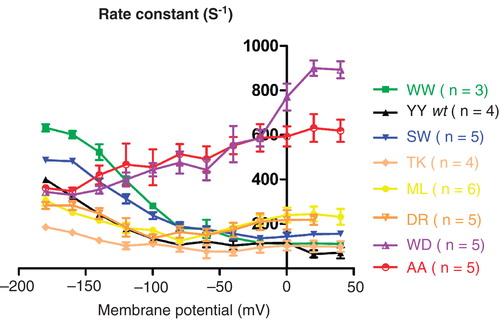
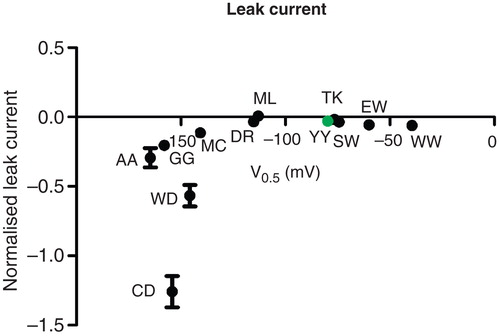
Figure 5. Leak versus sodium affinity. The inwardly rectifying leak current at −160 mV in a sodium containing, potassium-free extracellular environment compared to the affinity for extracellular sodium. The leak current was normalized to the potassium-induced pump current at +20 mV. This Figure is reproduced in colour in Molecular Membrane Biology online