Figures & data
Figure 1. Overview of structure prediction and docking pipeline applied to galanin receptors: Multiple sequence alignments of targets and templates; manual corrections of alignments based on predicted membrane topology and documented features of binding residues. The alignments are used for the creation of a large number of potential receptor models. Model filtering and selection of best models based on energy scoring function and cavity volume, docking of ligands, selection of best scoring poses and receptor-ligand contact analysis. This Figure is reproduced in colour in Molecular Membrane Biology online.
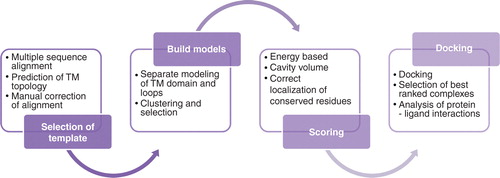
Table I. Sequence identity between galanin receptors and template alignments.
Table II. Amino acid residues involved in ligand binding to galanin receptor subtypes.
Figure 2. Multiple sequence alignment between the three galanin receptors and template structures (2R4R, 2RH1, 2ZIY). Rectangular boxes show the start and end residue positions for each transmembrane helix of GalR1, GalR2, and GalR3 subtypes. Convex lines connecting boxes denote the extracellular loops and the concave lines denote the intracellular loops. Colours highlight specific residues: in yellow – motifs characteristic to GPCR receptors; in blue – start and end of the transmembrane helices predicted by TMHMM; in red – start and end of the transmembrane helices in our models of the receptors; in green – overlap of residues in blue and red. Amino acids compared in are highlighted in purple; in italic are residues tested in site-directed mutagenesis studies (); in bold are residues in direct contact with ligands as found by docking (). This Figure is reproduced in colour in Molecular Membrane Biology online.
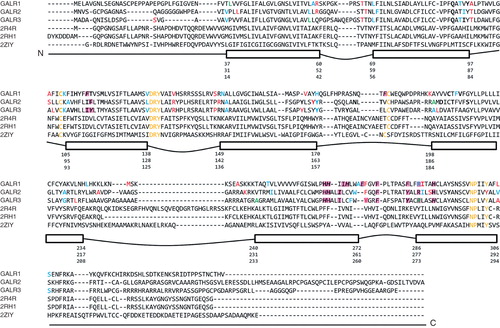
Table III. Cavity volumes and surface areas.
Table IV. Comparison of galanin receptors binding sites: Measuring spatial differences of residues positions corresponding in sequence and/or structural alignments. RMS calculated after molecular fit of TM backbone.
Table V. Selected ligand binding residues in galanin receptors found by docking studies.
Figure 3. Binding sites of galanin receptors subtypes with best binding ligands: (A) GalR1 and (B) GalR2 docked to galanin(2-6) and SNAP398299; GalR3 in complex with (C) two poses of galanin(2-6) and (D) two poses of SNAP398299. This Figure is reproduced in colour in Molecular Membrane Biology online.
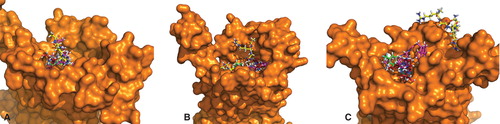
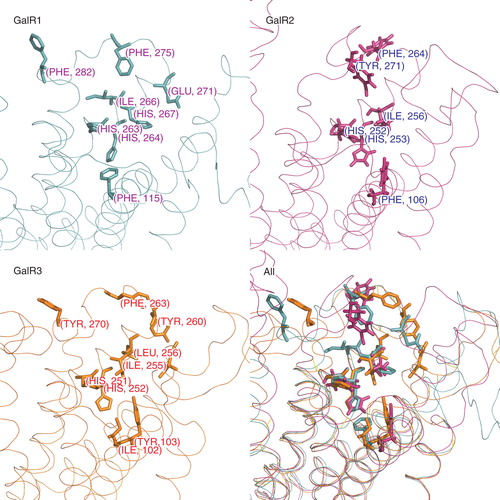
Figure 4. Amino acid residues in galanin receptors binding sites found in functional assays are shown separately for each receptor subtype (sticks). The last panel shows all structures superimposed. This Figure is reproduced in colour in Molecular Membrane Biology online.