Figures & data
Table 1. CTX-M ESBLs and their bacterial hosts.
Figure 1. Phylogenetic tree of CTX-M family based on amino-acid sequences. DNASIS Pro v2.10 (Hitachi Software Engineering Co., Tokyo, Japan) was used to align the amino-acid sequences and construct the phylogenetic tree. The amino-acid sequences were downloaded from GenBank under the accession numbers cited in . The branch lengths are drawn to scale and are proportional to the number of different amino-acid residues. The scale bars of 0.05 and 0.005 represent 5% and 0.5% amino-acid difference, respectively.
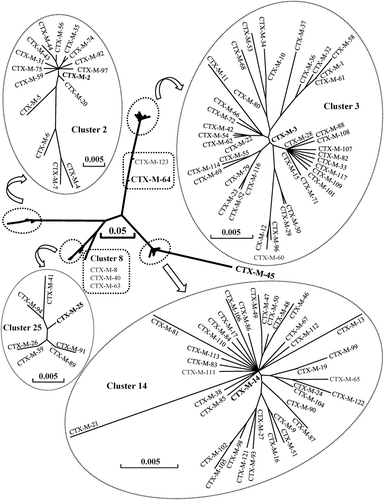
Table 2. Amino acid substitutions of CTX-M variants compared to their representative enzymes.
Figure 2. Comparison of amino-acid sequences of seven representative enzymes in the CTX-M family. Amino-acids are numbered according to the standard numbering scheme for the class A serine β-lactamases, giving the active site serine residue the Ambler number 70. Dots indicate identical amino-acids compared to CTX-M-2. Deletion mutations are expressed with short lines. The underlined amino-acids, 70SXXK73, 107P, 130SDN132, 143GG144, 166E and 234KXG236, represent the conserved residues in typical class A serine β-lactamases.

Figure 3. Identification of intrinsic cefotaximase genes in Kluyvera spp. as the original sources of acquired CTX-Ms based on their amino-acid identities and the homologies of neighboring sequences of the associated genes. c-CTX-M, CTX-M identified on chromosome of Kluyvera spp.; p-KLUC-2, KLUC-2 identified on plasmid in a clinical isolate of Enterobacter cloacae.
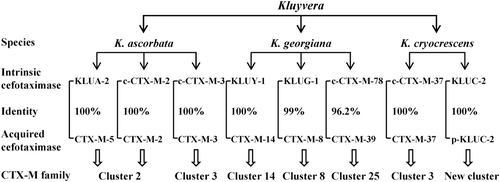
Table 3. Genetic platforms of CTX-M enzymes.
Figure 4. Typical genetic platforms of CTX-M enzymes. A & B: the blaCTX-M gene cassettes bracketed upstream by ISEcp1/ISEcp1-like and downstream by IS903/IS903-like (A) or orf477/orf477-like (B); C: blaCTX-M genes associated with class 1 integron-ISEcp1; D & E: blaCTX-M genes associated with class 1 integron-ISCR1 complex. CS, conserved segment; intI, integrase gene; qacE▵1, quaternary ammonium resistance gene; sul1, sulphonamide resistance gene; 3′-CS2, the second copy of 3′-conserved segment.
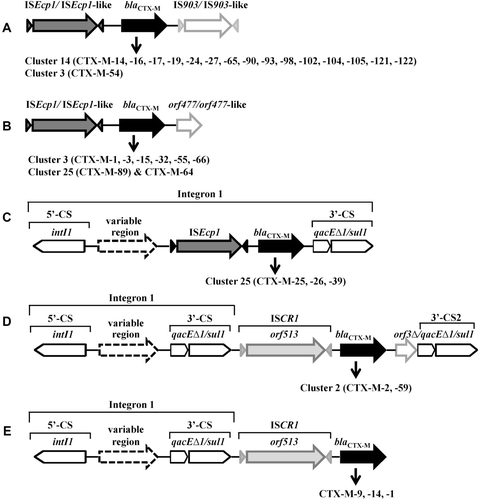
Table 4. Plasmids associated with the spread of CTX-M genes.
