Figures & data
Figure 1. Simplified scheme of PMCA structure with sites modified by high glucose. Transmembrane regions are numbered 1–10. Approximate positions of the phosphoryl-aspartate (D*) and lysine (K) crucial for ATP binding are shown. In the C-terminal region, tyrosine (Y) residue is indicated. Its phosphorylation results in inactivation of the pump. Positions of particular residues are numbered according to the human PMCA4b sequence.
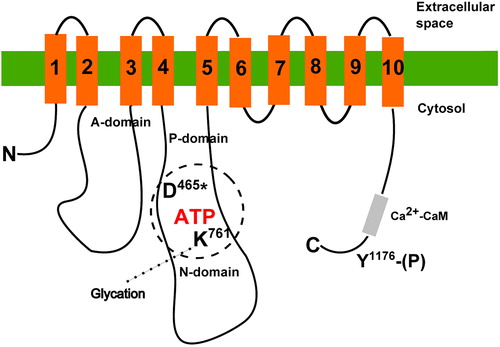
Figure 2. Structures of noncrosslinking and crosslinking AGEs in Ca2+-pumps. Formation of advanced glycation end-products through glucose-protein Schiff base and Amadori rearrangement (a), and structures of noncrosslinking (b) and crosslinking (c) AGE molecules most frequently formed on Ca2+-pumps.
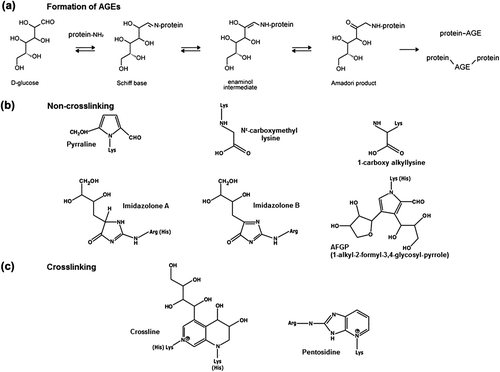
Figure 3. Planar model of the primary and estimated secondary structure of rat SERCA2b isoforms highlighting AGEs formation and posttranslational modifications. The model is adapted from Dode et al. [Citation108] and based on the crystal structure of SERCA1. Each circle corresponds to an amino acid residue. Amino acids arginines R164, 636, lysines K476, 481, and histidine H526 are targets for noncrosslinking AGEs like imidazolone A, imidazolone B and AFGP (red circles with black letters). Between amino acids K135 and R164, K141 and R677, K135 and R655, K460 and R636, and between K683 and R619 crosslinking AGEs-like pentosidine (dotted red circles with red letters) are formed. Tyrosines Y294 and Y295 are documented sites for nitration (presented by pink circles and pink letters). Cysteine C674 is glutathionylated in the presence of NO (blue letters in blue circles). Asparagine N1035 is a potential glycosylation target (orange circle with orange letter). Aspartic acid D351 (black circle and letter with the phosphate group indicated) represents the phosphorylation site. The nucleotide-binding domain spans from residue T357 to L599 (shown as green circles with white letter) and the most crucial amino acid residues for ATP binding are lysines K514, K492 and phenylalanine F487 (green circles with black letters). Amino acid residues contributing to Ca2+ ligand binding are shown as black circles with black letters in the high affinity Ca2+ binding site of the transmembrane domain on M4, M5, M6, and M8 helixes.
![Figure 3. Planar model of the primary and estimated secondary structure of rat SERCA2b isoforms highlighting AGEs formation and posttranslational modifications. The model is adapted from Dode et al. [Citation108] and based on the crystal structure of SERCA1. Each circle corresponds to an amino acid residue. Amino acids arginines R164, 636, lysines K476, 481, and histidine H526 are targets for noncrosslinking AGEs like imidazolone A, imidazolone B and AFGP (red circles with black letters). Between amino acids K135 and R164, K141 and R677, K135 and R655, K460 and R636, and between K683 and R619 crosslinking AGEs-like pentosidine (dotted red circles with red letters) are formed. Tyrosines Y294 and Y295 are documented sites for nitration (presented by pink circles and pink letters). Cysteine C674 is glutathionylated in the presence of NO (blue letters in blue circles). Asparagine N1035 is a potential glycosylation target (orange circle with orange letter). Aspartic acid D351 (black circle and letter with the phosphate group indicated) represents the phosphorylation site. The nucleotide-binding domain spans from residue T357 to L599 (shown as green circles with white letter) and the most crucial amino acid residues for ATP binding are lysines K514, K492 and phenylalanine F487 (green circles with black letters). Amino acid residues contributing to Ca2+ ligand binding are shown as black circles with black letters in the high affinity Ca2+ binding site of the transmembrane domain on M4, M5, M6, and M8 helixes.](/cms/asset/51aad77e-0644-4cdf-b78d-067facb5b73a/ifra_a_807923_f0003_b.jpg)
