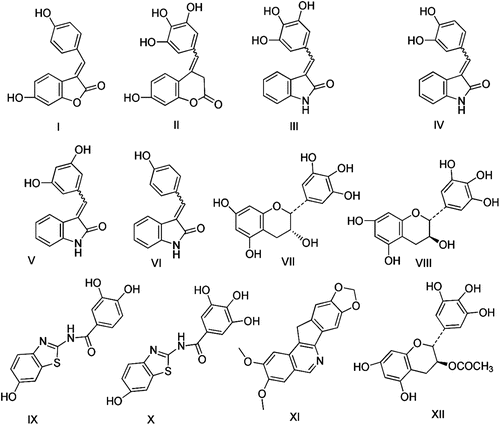
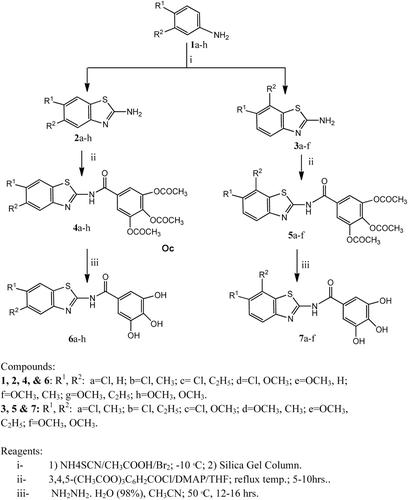
Figures & data
Table 1. Simulation fitting scores of the designed triacetyl (4a-h and 4a-f) and the free hydroxyl derivatives (6a-h and 7a-f) with the generated ideal hypothesis of topoisomerase I inhibitors in comparison to the leads compounds; I-XII.
Table 2. Docking energy and binding pattern of the docked triacetyl (4a-h and 5a-f) and the free hydroxyl derivatives (6a-h and 7a-f) with topo-I enzyme in comparison to the lead XI: (numbers of HB, numbers of Wan Der Wall attractions (pi-pi and sigma-pi) interactions and [pi-(+) interaction distance with Arg364 at the binding sites).
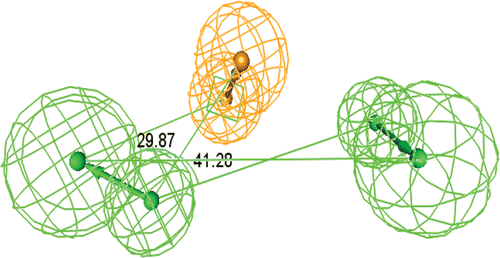
Figure 2. Constraint distances in the ideal Hypothesis of Topoisomerase-I inhibitors. (HBA = Hydrogen bond acceptor feature, AR =Ring Aromatic vector feature).
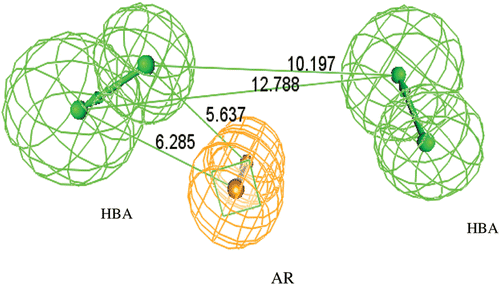
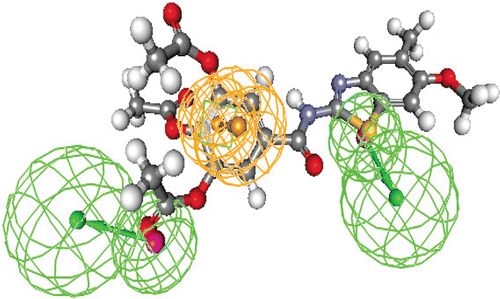
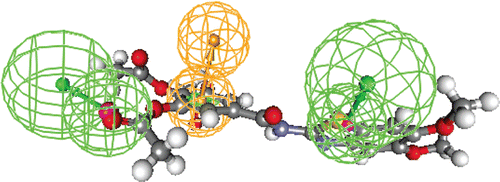
Figure 6. Publication quality 3D diagram of the X-ray crystal structure of lead (XI)complexed with topoisomerase-I enzyme (redocked with RMSD = 0.0023) and Docking value = −44.41. The binding pattern showed: [1HB with Arg364 (as reportedCitation27 and 8 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [ pi-(+)] interaction between it and Arg364 by distance = 6.95 Å.
![Figure 6. Publication quality 3D diagram of the X-ray crystal structure of lead (XI)complexed with topoisomerase-I enzyme (redocked with RMSD = 0.0023) and Docking value = −44.41. The binding pattern showed: [1HB with Arg364 (as reportedCitation27 and 8 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [ pi-(+)] interaction between it and Arg364 by distance = 6.95 Å.](/cms/asset/85f0f9ba-061d-4397-b843-824bfe8f5e7d/ienz_a_716835_f0007_b.gif)
Figure 7. Publication quality of the top docked compound 5f with topoisomerase-I enzyme with Docking value = −67.55. The binding pattern showed: [5 HB with the surrounded amino acids and DNA nucleotides including Arg364 (as crucial SAR), 7 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [pi-(+)] interaction between it and Arg364 by distance =4.49 Å].
![Figure 7. Publication quality of the top docked compound 5f with topoisomerase-I enzyme with Docking value = −67.55. The binding pattern showed: [5 HB with the surrounded amino acids and DNA nucleotides including Arg364 (as crucial SAR), 7 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [pi-(+)] interaction between it and Arg364 by distance =4.49 Å].](/cms/asset/3f3dcf3b-f7c9-40a8-95b4-47b8eae6caf9/ienz_a_716835_f0008_b.gif)
Figure 8. Publication quality of the lowest docked compound 6b with topoisomerase-I enzyme with Docking value = −45.10. The binding pattern showed: [3HB with the surrounded amino acids and DNA nucleotides including Arg364 (as a crucial SAR), 6 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [ pi-(+)] interaction between it and Arg364 by distance =5.69 Å].
![Figure 8. Publication quality of the lowest docked compound 6b with topoisomerase-I enzyme with Docking value = −45.10. The binding pattern showed: [3HB with the surrounded amino acids and DNA nucleotides including Arg364 (as a crucial SAR), 6 lipophilic pi-pi interactions with the surrounded amino acids and DNA nucleotides, and a [ pi-(+)] interaction between it and Arg364 by distance =5.69 Å].](/cms/asset/1ff90026-d09f-4768-bf32-9141fe92ff41/ienz_a_716835_f0009_b.gif)
Table 3. Effects of test set compounds (4a~7f) and the reference drugs (doxorubicin and camptothecin) toward the growth of human cervix carcinoma (Hela) and human breast carcinoma (MCF7) cell lines expressed in IC50 values, % Relative IC50 and numbers of folds of more activity than the given reference drugs.