Figures & data
Table 1. Human (h) hCA I, II and yeast (S. cerevisiae) ScCA inhibition data with phenols 1–13 by a stopped flow CO2 hydrase assayCitation31.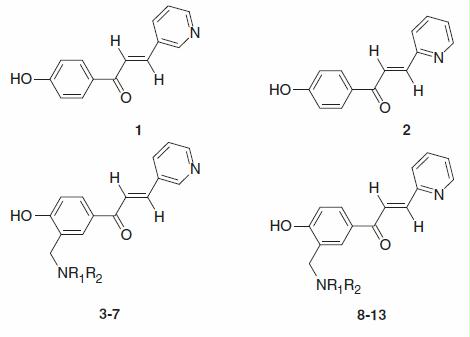
Figure 1. Alignment of the amino acid sequences of selected β-CAs from four species. The S. cerevisiae numbering system was used. Zinc ligands (amino acids: 57, 112 and 115) are indicated in red whereas the catalytic dyad (amino acids 59 and 61) typical of β-CAs in blue. The asterisk (*) indicates identity at a position; the symbol (:) designates conserved substitutions, while (·) indicates semi-conserved substitutions. Multiple alignment was performed with the program Clustal W, version 2.1 (http://www.ebi.ac.uk/services/proteins). Sequence accession numbers are indicated in .

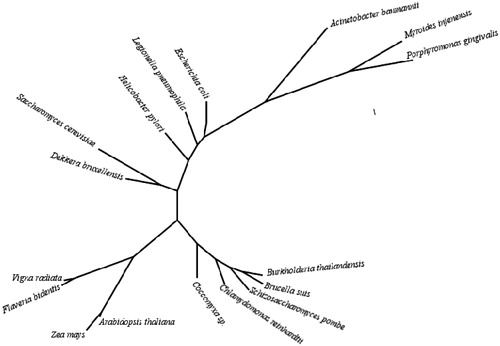
Figure 2. Phylogenetic tree of the beta CAs from selected eukaryotic and prokaryotic species. The tree was constructed using the program PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/). Organisms and sequence accession numbers are as follows: Schizosaccharomyces pombe (CAA21790); Brucella suis 1330 (NP_699962.19); Burkholderia thailandensis Bt4 (ZP_02386321); Coccomyxa sp. (AAC33484.1); Chlamydomonas reinhardtii (XP_001699151.1); Acinetobacter baumannii (YP_002326524); Porphyromonas gingivalis ATCC 33277 (YP_001929649.1); Myroides injenensis (ZP_10784819); Zea mays (NP_001147846.1); Vigna radiata (AAD27876); Flaveria bidentis, isoform I (AAA86939.2); Arabidopsis thaliana (AAA50156); Helicobacter pylori (BAF34127.1); Legionella pneumophila 2300/99 (YP_003619232); Escherichia coli (ACI70660); Saccharomyces cerevisiae (GAA26059); Dekkera bruxellensis AWRI1499 (EIF49256).