Figures & data
Figure 1. Generic chemical structure of the three classes of compounds assayed in this work: (E)-chalcones 1–82, (E)-N′-benzylidene-benzohydrazides 83–99, and alkyl-esters of gallic acid 100–110.
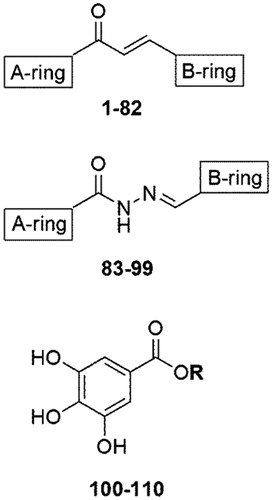
Table 1. IC50 values of compounds active against recombinant falcipain-2.
Figure 2. Lineweaver–Burk plots of the kinetic behavior of inhibitors 48, 54 and 66. The plots represent an average of at least five different experiments.
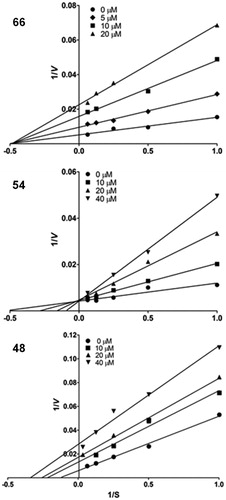
Figure 3. Peptide mass fingerprint of falcipain-2 with a coverage of 73%. Inset: peak m/z 2263.324 corresponds to the peptide-containing the catalytic Cys42. The spectrum represents a sum of 4000 lasers shots subtracted from the baseline.
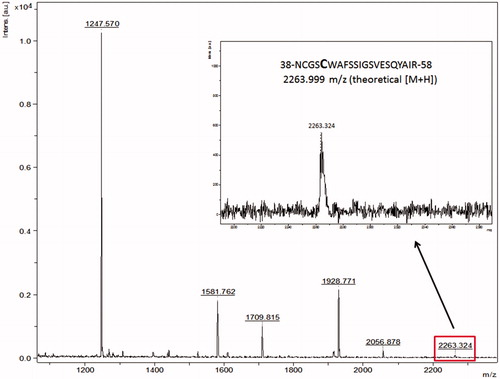
Figure 4. Peptide mass fingerprint of falcipain-2 in the presence of the three best inhibitors. The arrows indicate the peptide-containing the catalytic Cys42, m/z 2263.324. Panels show falcipain-2 without inhibitors (A), in the presence of the non-competitive inhibitor 66 (B), in the presence of the competitive inhibitor 54 (C) and in the presence of the mixed-type inhibitor 48 (D). The spectrum represents a sum of 4000 lasers shots subtracted from the baseline.
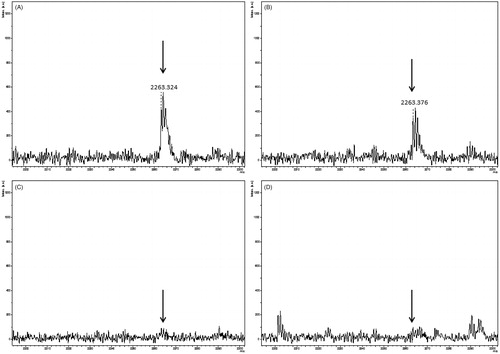
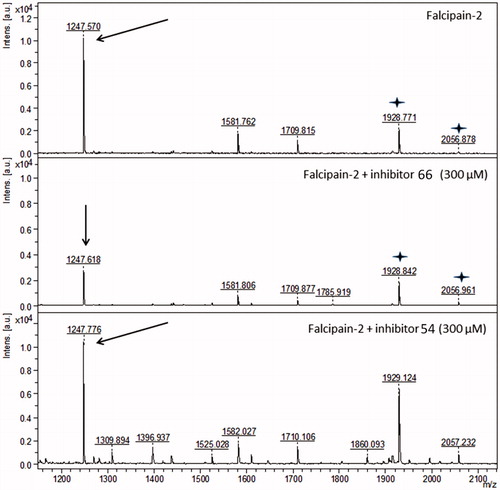
Figure 5. Peptide mass fingerprint of falcipain-2. The peak corresponding to the peptide 204-NSWGQQWGER-213 is labeled (m/z 1247.719, arrows). The panels show falcipain-2 in the absence of the inhibitors, in the presence of the non-competitive inhibitor 66, and in the presence of the competitive inhibitor 54. The spectra represents a sum of 4000 lasers shots subtracted from the baseline. Stars represent the peaks m/z 1928.771 and m/z 2056.878, which are not affected by inhibitor 66.