Figures & data
Table 1. Effect of the N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives on in vitro platelet aggregation induced by arachidonic acid (500 μM); collagen (5 μg/mL); ADP (3 μM) and epinephrine (2 μM).
Table 2. Comparison of stereoelectronic parameters of the N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives and their interaction with feasible targets, cyclooxygenase-1 (COX-1) and thromboxane synthase (TXS), structure–activity relationship of derivatives.
Table 3. Effect of N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives on in vitro plasma TXB2 and PGH2 level determination assays.
Figure 1. No active inhibition of the coagulation cascade is observed for N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives in both extrinsic (A) or intrinsic pathways (B) determined by PT and aPTT assays using human platelet-poor plasma, (C) effect on healthy human erythrocytes. Pyrazolopyridines (100 μM) incubated with suspension of erythrocytes in 37 °C at different times show no significant hemolysis (>10%). Triton X-100® was used as positive control and DMSO 1% as negative control. The assays were performed in triplicate in two independent experiments. * % hemolysis >10%.
![Figure 1. No active inhibition of the coagulation cascade is observed for N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives in both extrinsic (A) or intrinsic pathways (B) determined by PT and aPTT assays using human platelet-poor plasma, (C) effect on healthy human erythrocytes. Pyrazolopyridines (100 μM) incubated with suspension of erythrocytes in 37 °C at different times show no significant hemolysis (>10%). Triton X-100® was used as positive control and DMSO 1% as negative control. The assays were performed in triplicate in two independent experiments. * % hemolysis >10%.](/cms/asset/64e31b4b-ebff-479a-a9ef-f6a978e5f8ec/ienz_a_1158712_f0001_b.jpg)
Figure 2. (A) Chemical structure and substituents of N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives (3a, 3b, 3d, 3h and 3j). (B) Electrostatic potential map of N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives. The black square marks the R2 position. Docking complexes are observed between derivatives 3a (C), 3b (D) and 3h (E) with thromboxane synthase. π–cation interactions with the heme iron are shown in purple. Hydrogen bonds are shown as dashed orange lines.
![Figure 2. (A) Chemical structure and substituents of N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives (3a, 3b, 3d, 3h and 3j). (B) Electrostatic potential map of N′-substituted-phenylmethylene-3-methyl-1,6-diphenyl-1H-pyrazolo[3,4-b]pyridine-4-carbohydrazide derivatives. The black square marks the R2 position. Docking complexes are observed between derivatives 3a (C), 3b (D) and 3h (E) with thromboxane synthase. π–cation interactions with the heme iron are shown in purple. Hydrogen bonds are shown as dashed orange lines.](/cms/asset/cb8e4071-2df6-4781-8acc-fd78d0ae8da1/ienz_a_1158712_f0002_c.jpg)
Figure 3. (A) Survival rates of mice treated with pyrazolopyridine derivatives on in vivo pulmonary thromboembolism induced by thrombin. (B) Effects of pyrazolopyridine treatment on total blood loss through tail bleeding assay. (C) Monitoring of total hemoglobin loss from animals treated with each derivative over a course of 30 min. (D) Ex vivo recalcification time of PRP isolated from mice treated with each derivative. All derivatives and ASA were assayed at 10 mM/kg and RIVA (Rivaroxaban) at 1 mM/kg. The assays were performed with three groups of three animals for each compound or control. The results are presented as the mean ± SD. *p <0.05.
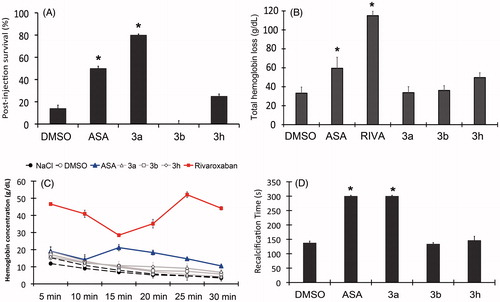
Figure 4. Histopathology of the lungs isolated from mice subjects included for in vivo pulmonary thromboembolism (HnE staining, 8× magnification; scale bar = 600 μm). Microscopic emboli (*) are seen for all treated groups. (A–D) Intense bronchoconstriction is seen for all treated groups with a slight integrity of alveoli (•) observed for the 3a-treated group, which also displays increased blood perfusion (▪). Only derivative 3a was used because the high survival rate was observed previously (D). All derivatives and ASA were assayed at 10 mM/kg (HnE staining, 8× magnification; scale bar = 600 μm).
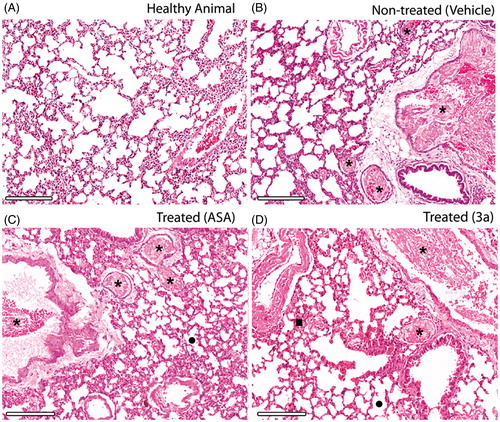
Figure 5. Histopathology of the lungs of thrombin-induced animals at the same day (A) and five days after induction (B). Integrity of alveoli and increased blood perfusion are similar for both, but microscopic emboli (*) after five days from the induction show a residual thrombi starting to disassemble. 3a derivative at 10 mM/kg (HnE staining, 8× magnification; scale bar = 600 μm).
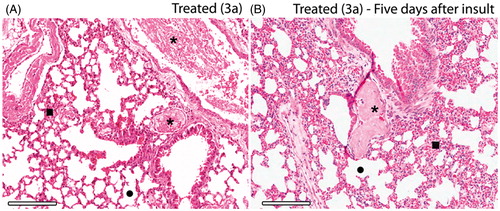
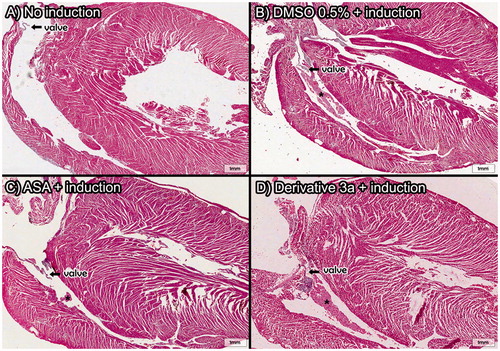
Figure 6. Heart histopathology after in vivo pulmonary thromboembolism model assays induced by thrombin. Thrombi located in the right ventricle reveal the migration of microemboli under formation to the lungs of the animal subjects. Only derivative 3a was used because the high survival rate was observed previously. All derivatives and ASA were assayed at 10 mM/kg (HnE staining, 8× magnification; scale bar = 600 μm) (A) Control group and *=thrombi.