Figures & data
Figure 1. Cross-reactivity of PTPμ intracellular antibodies with other type IIb RPTPs. Sf9 cells infected to express the type IIb RPTPs subfamily members were lysed and analyzed by immunoblotting with the following anti-PTPμ cytoplasmic antibodies: SK6, SK7, SK10, SK15, SK18, and a C-terminal PTPμ antibody (A). Proteins expressed at the cell surface were isolated and immunoblotted with SK7 and SK18 (B). A summary of the cross-reactivity of the SK series of antibodies and the C-terminal PTPμ antibody with type IIb RPTP members is shown in (C).
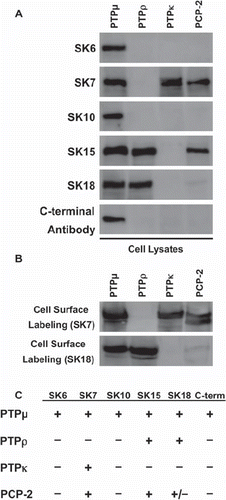
Figure 2. Binding of the SK series of PTPμ antibodies is eliminated when specific amino acids are deleted in the cytoplasmic domain of PTPμ fusion proteins. Recombinant PTPμ fusion proteins containing discrete amino acid composition were resolved by SDS-PAGE and immunoblotted with the SK series of antibodies and the C-terminal PTPμ antibody (A). An illustration of the PTPμ recombinant proteins is shown in (B) along with a summary of the epitope binding experiments.
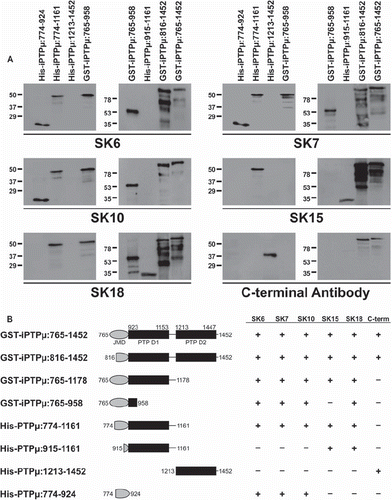
Figure 3. Identification of the epitopes of the SK series of PTPμ antibodies based on amino acid sequence comparison between type IIb RPTPs. SK6 and SK10 (A) are predicted to bind to amino acids 816–914 in the juxtamembrane domain (JMD-W) and are outlined by a black box. SK7 is predicted to bind to the residues outlined by the black box in (B), 850–855 or 868–877, also in JMD-W. SK15 likely recognizes 958–1161 in the first phosphatase domain (PTP1) (C), whereas SK18 recognizes the residues outlined by the black box in (D), 941–947, also in PTP1. The PTPμ C-terminal antibody is known to bind to residues 1436–1452 (E). Regions of identity between the type IIb RPTP amino acid sequences are shaded gray.
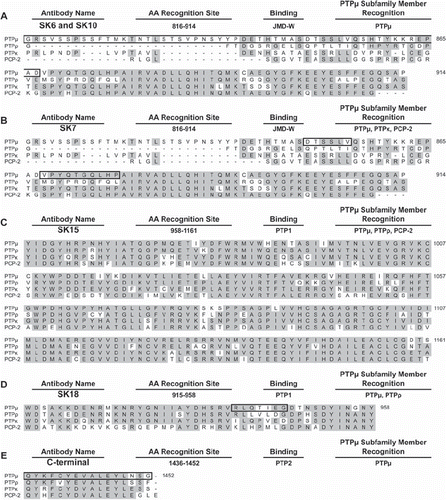
Figure 4. PCP-2 does not mediate homophilic cell-cell aggregation. Nonadhesive Sf9 cells were infected with baculoviruses expressing different type IIb RPTP subfamily members and allowed to aggregate for 30 min prior to imaging. PTPμ, PTPρ, PTPκ all mediate homophilic cell aggregation, whereas PCP-2 does not.
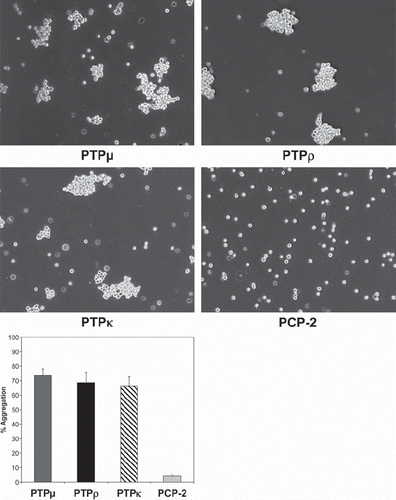
Figure 5. PCP-2 does not interact in trans with other type IIb RPTPs to mediate heterophilic cell-cell aggregation. Sf9 cells expressing both GFP and PCP-2 were mixed with Sf9 cells expressing RFP and either PTPμ, PTPρ, or PTPκ. After 30 min of aggregation, the cells were imaged. Phase-contrast images show that aggregates form. The overlay fluorescent images demonstrate that these aggregates are composed exclusively of RFP-expressing cells, and exclude GFP/PCP-2–expressing cells.
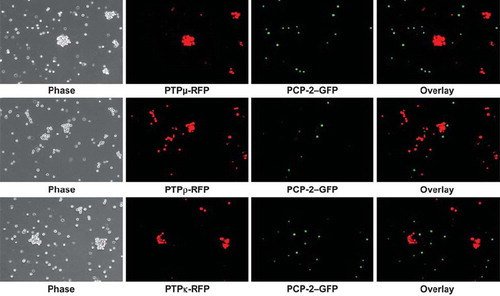
Figure 6. PTPκ-expressing cells sort away from PTPμ- and PTPρ-expressing cells. Sf9 cells coinfected with RFP and PTPκ were mixed with cells coinfected with GFP and either PTPμ or PTPρ and subjected to aggregation assay for 30 min. A control experiment with Sf9 cells expressing only RFP or GFP demonstrate that these two fluorescent proteins do not induce Sf9 cell aggregation. The overlay images demonstrate that only exclusively red or green aggregates formed in all aggregation experiments, demonstrating that the type IIb RPTPs sort away from each other. Phosphatase expression was detected in Sf9 cells grown in culture by immunocytochemistry with SK15 antibody and a Texas Red secondary antibody. The GFP expression following coinfection is shown in green. The overlay image demonstrates that GFP is coexpressed in the same cells as the phosphatases PCP-2 or PTPμ.
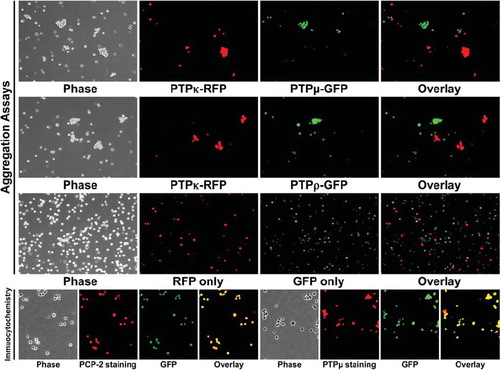
Figure 7. Expression of PTPρ-PCP-2 chimeric proteins on the surface of Sf9 cells. PTPρ-PCP-2 chimera proteins were engineered (A) and expressed in Sf9 cells. Within the extracellular segment, the oval represents the MAM domain, the circle with the S-S symbol represents the Ig domain, and the solid rectangles represent the FNIII repeats. In the intracellular segment, the oval represents the juxtamembrane domain, and the aqua shaded rectangles represent the PTP domains. Total cellular protein as well as cell surface expressed proteins were isolated and immunoblotted with SK15 (B). Control expression of actin is shown for the cell lysates (B). There is no actin contamination of the cell surface, labeling samples thus serving as a control for the procedure.
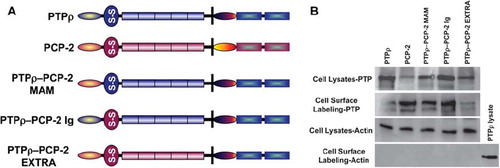
Figure 8. Swapping of either the MAM or Ig domain of PCP-2 into PTPρ creates a functional protein capable of mediating weak cell-cell aggregation. PTPρ-PCP-2 MAM and PTPρ-PCP-2 Ig chimeric proteins are both capable of mediating weak cell-cell aggregation (A), more than PCP-2 but less than wild-type PTPρ (B). PTPρ-PCP-2 Extra is not capable of mediating cell-cell aggregation.
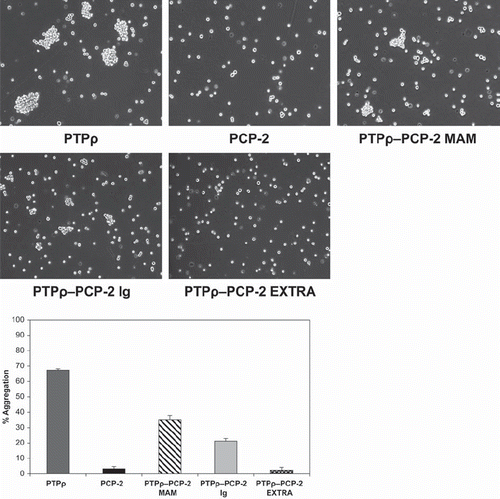
Figure 9. Chimeric PTPρ-PCP-2 proteins sort away from each other. Sf9 cells coinfected with RFP and PTPρ were mixed with cells coinfected with GFP and the chimeric PCP-2 proteins and evaluated in aggregation assays for 30 min. The overlay images demonstrate that only exclusively red or green aggregates formed in these experiments, demonstrating that chimeric proteins sort away from wild-type PTPρ and create a unique recognition site. A summary table of all heterophilic binding experiments is shown at the bottom of this figure.
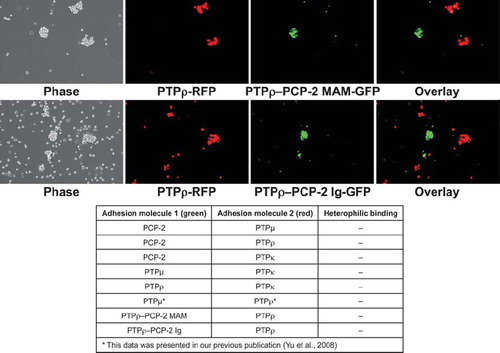
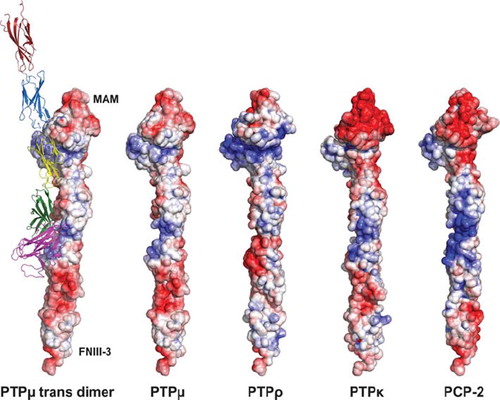
Figure 10. Representation of type IIb RPTPs using solvent accessible surface charge data. The diagram is contoured from −10 kT to 10 kT with a red (negative electrostatic potential) to white (neutral charge) to blue (positive electrostatic potential) ramping scheme. The location of the MAM and FNIII-3 domains are indicated to orient the reader. The ribbon diagram illustrates both the trans-dimer interface and the various domains described in this article. The MAM domain is shown as magenta. The Ig domain is shown in green. FNIII repeat 1 is yellow, whereas FNIII repeat 2 is blue and FNIII repeat 3 is red.