Figures & data
Figure 1. Mass spectra showing compounds of IM-133N extract. Spectrometric conditions are described in the Methods.
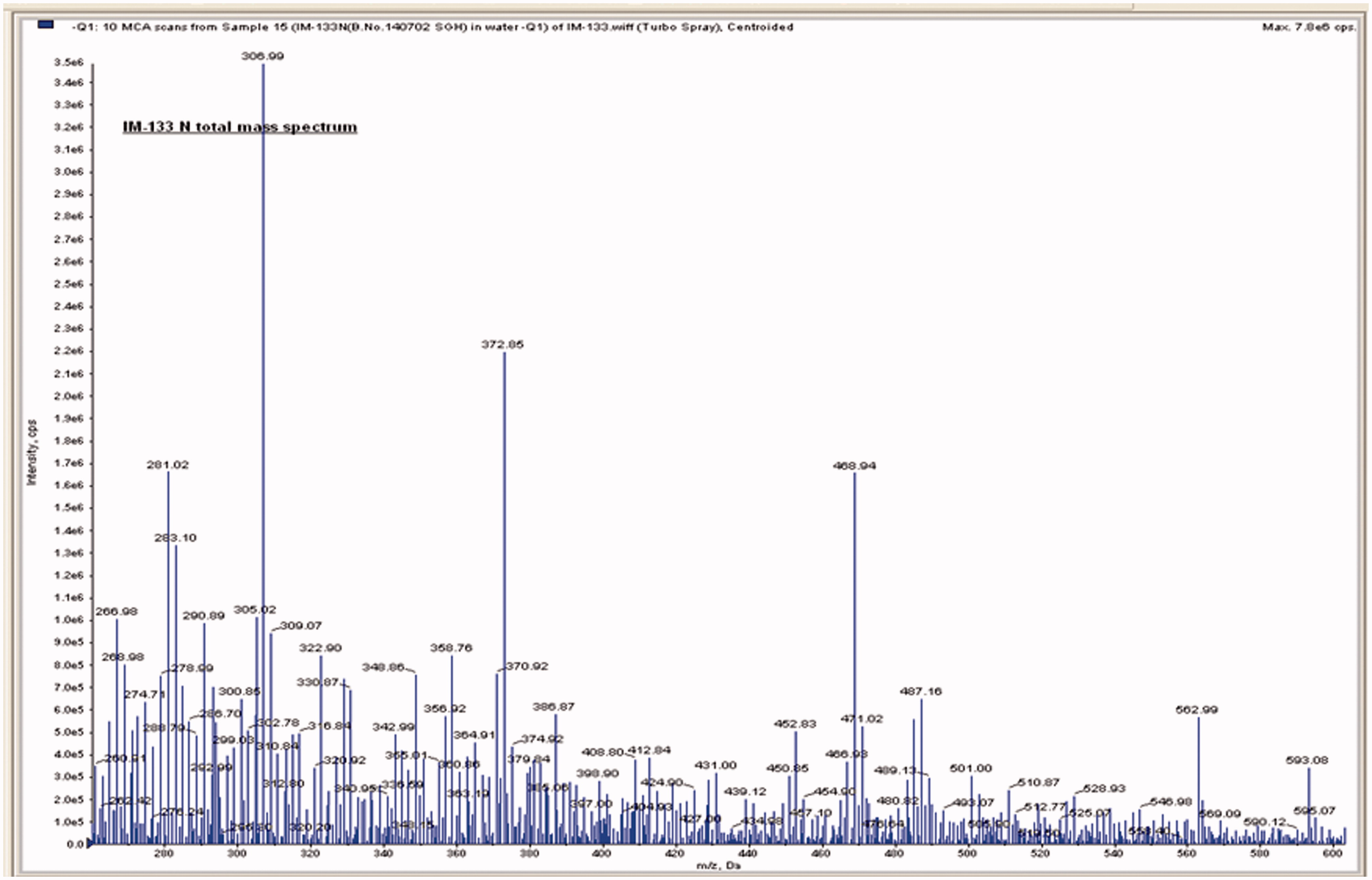
Table 1. List of compounds in IM-133N identified by mass spectrometry.
Table 2. Primers used for amplifications.
Figure 2. Cytotoxicity of IM-133N on (a) RAW264.7 and (b) THP-1 cells. Cells were incubated for 24 h with differing concentrations of IM-133N and cell viability was then determined using an MTT assay. Data are expressed as mean (±SE) cytotoxicity (i.e. percentage reduction in viable cell number vs control) from three assays using n = 3 samples/IM-133N concentration.
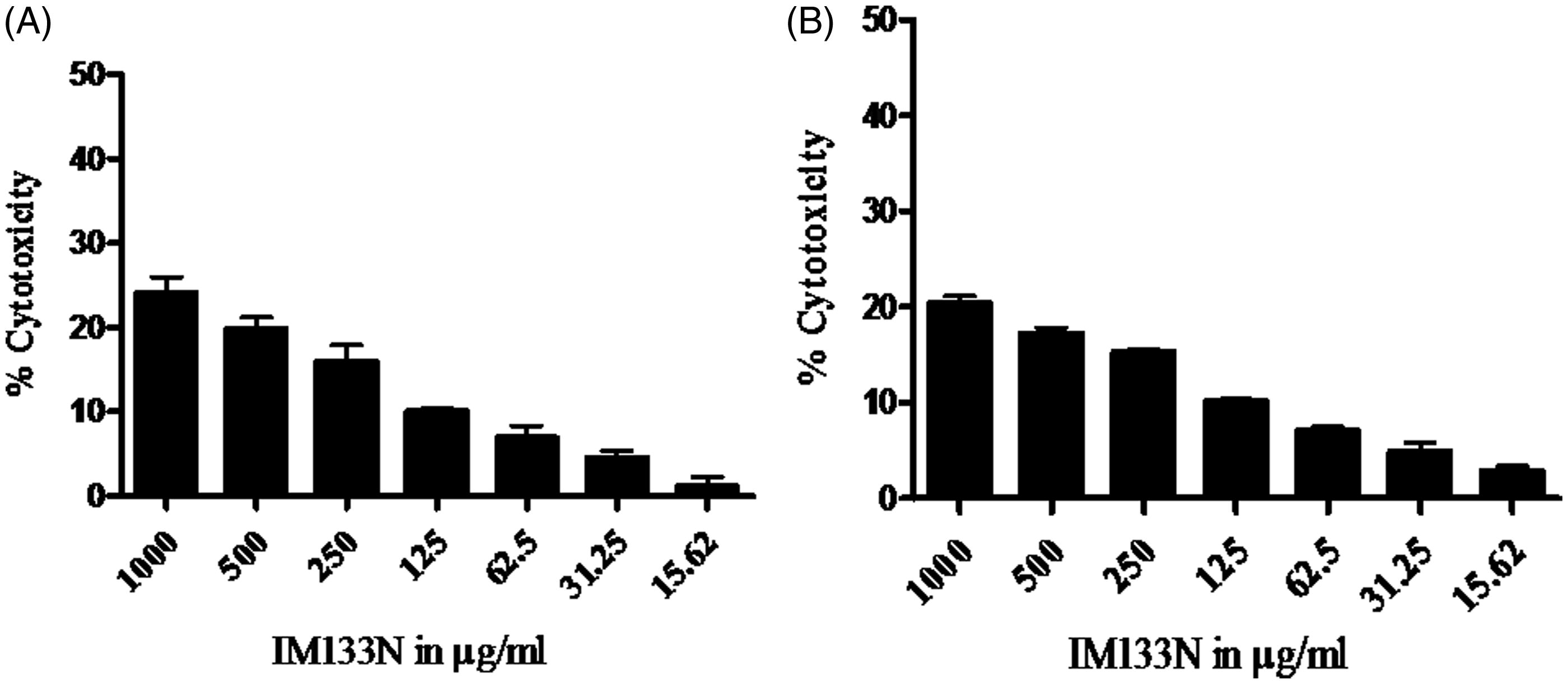
Table 3. Effect of IM133N on NO and TNFα secretion.
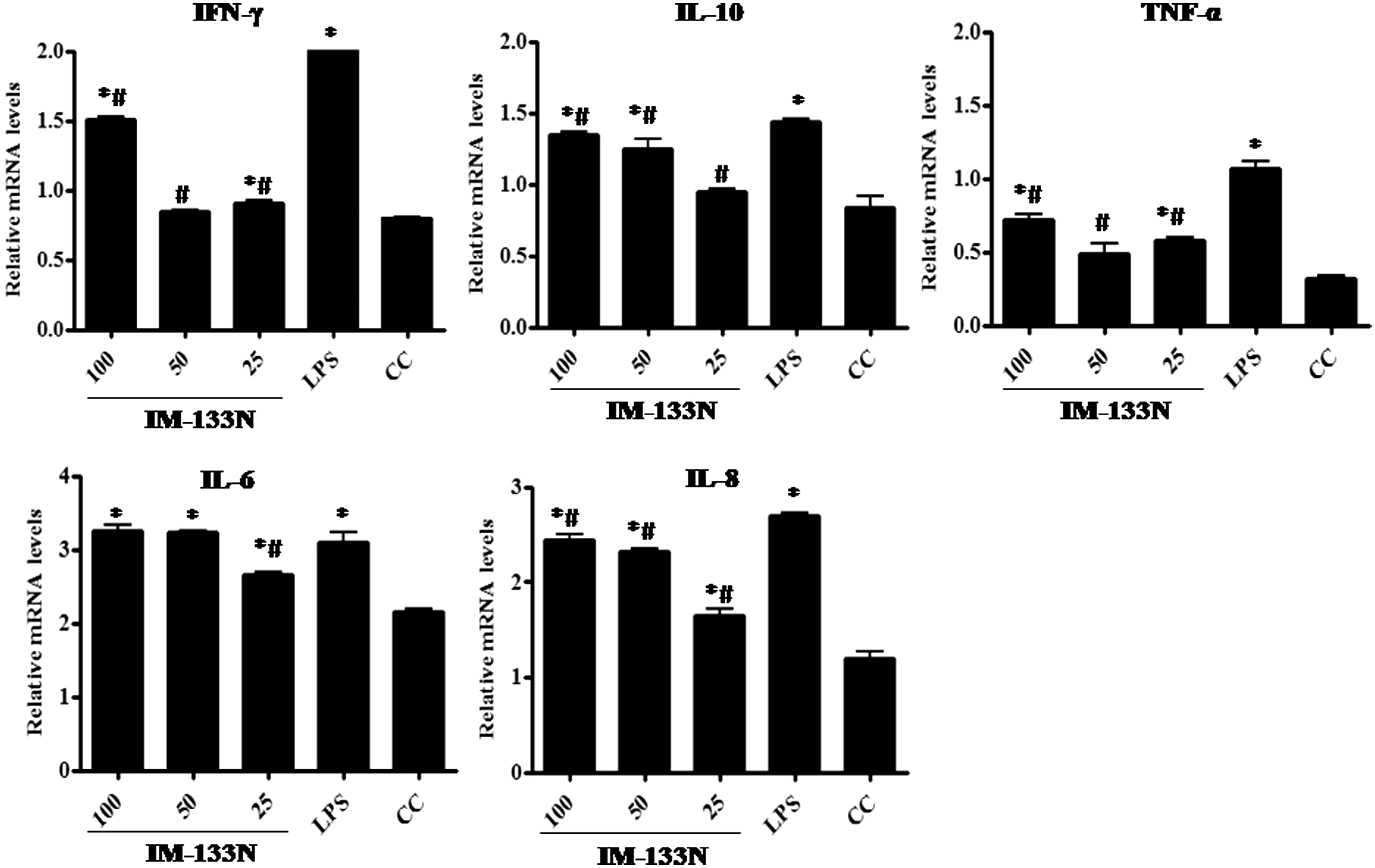
Figure 3. Densitometric analysis of gene transcripts obtained from RAW264.7 cells. Cells were incubated with the IM-133N for 4 h (for TNFα gene amplification) or for 24 h (for iNOS and the other cytokines investigated) before being harvested. Relative levels of each gene’s expression were normalized to that for GAPDH. Values shown depict arbitrary units. Values are mean ± SD of three individual experiments using n = 3 samples/IM-133N concentration. *Value significantly different from control or # from LPS control (p < 0.05).
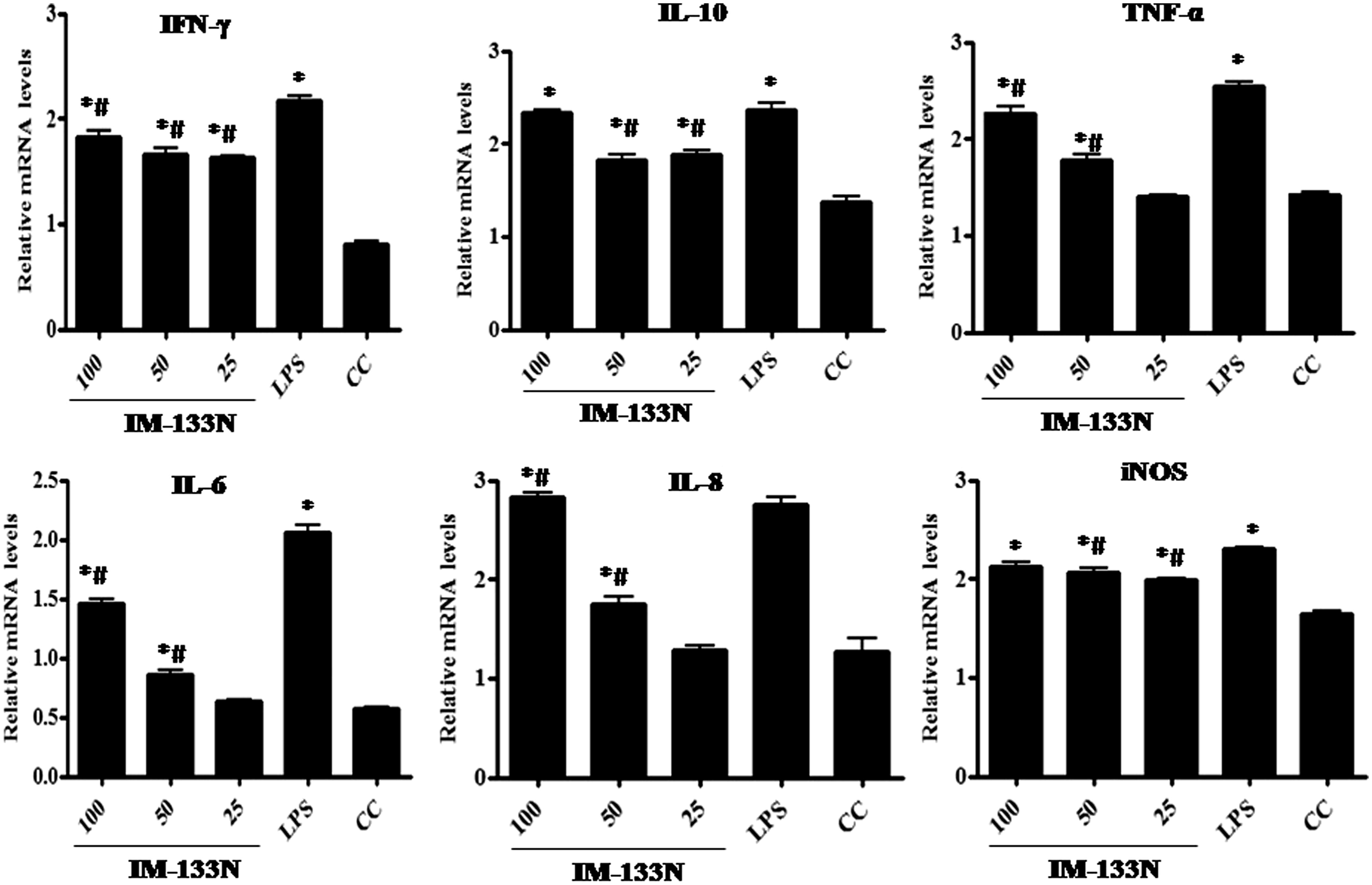
Figure 4. Densitometric analysis of gene transcripts obtained from THP-1 cells. Cells were incubated with the IM-133N for 4 h (for TNFα gene amplification) or for 24 h (for the other cytokines investigated) before being harvested. Relative levels of each gene’s expression were normalized to that for GAPDH. Values shown depict arbitrary units. Values are mean ± SD of three individual experiments using n = 3 samples/IM-133N concentration. * Value significantly different from control or # from LPS control (p < 0.05).