Figures & data
Figure 1. Schematic representation of PRM1 and PRM2 gene nucleotide sequence. The blue colored regions are introns. Nucleotide changes are green and the variant is shown just above in brown with their respective position. The corresponding amino acid change is shown below the wild type in red.

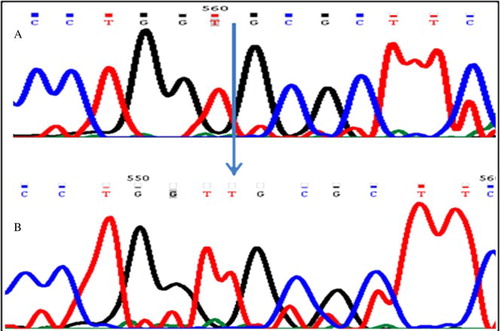
Figure 2. Sequence electrophoreogram of the SNP variant. The nucleotide variant SNP c443C > A in PRM2 gene is resolved. A) represents wild type sequence (C/C); B) represents sequence showing homozygous mutant (A/A).
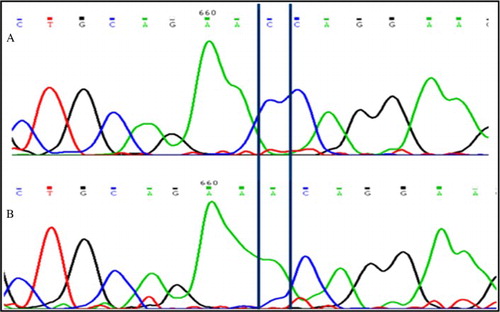
Table 1. Sperm PRM and TNP genotypic and allelic frequency in oligozoospermic infertile and control men.
Figure 3. Schematic representations of TNP1 and TNP2 genes showing the location of the SNPs. Blue-colored regions are introns. Nucleotide changes are green and the variant is shown just above in brown with their respective position. The corresponding amino acid change is shown below the wild type in red. The non coding exon region is shown underlined.
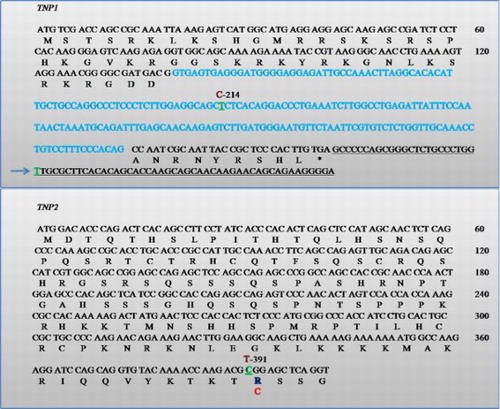
Figure 4. Sequence electrophoreogram of the SNP variant. The insertion c. Ins T 396-397 3′UTR TNP1 gene is highlighted. A) represents sequence showing homozygous wild-type (GGTG); B) represents sequence showing insertion (GGTTG).