Figures & data
Figure 1. Differentiation of human embryonic stem cells (hESCs) into adipocytes and the identification of adipocytes. (A) Thawed clones before passage ×100. The thawed hESCs are incubated in an inactivated mixed feeder layer (1:1 of mouse embryonic fibroblasts: human foreskin fibroblasts) at 37°C in an atmosphere of 5% CO2 for two days. Next day, hESCs are observed under an inverted microscope. hESCs grow in colony, and are flat in shape with clear cell boundaries. (B) Adipocytes induced for 21 days ×40. Three days after induction, clones become loose with irregular edge. Five days after induction, lipid droplets are seen in cytoplasm with the nucelus in cellular center. Two weeks after induction, mature adipocytes occur. More and more lipid droplets gather, and then cells become round or oval in shape and the nucelus are pulled to one side of the cells. Differentiation peak occurs 21 after induction. (C) Oil red O staining ×40. After hESCs differentiate into mature adipocytes, cells become round with strong refractivity and lipid droplets gather. Lipid droplets in mature adipocytes can be specifically stained with oil red O, showing red color. (D) peroxisome proliferator-activated receptor gamma (PPARγ)2 mRNA expression detected by RT-PCR RNA in cells is extracted with Trizol, and then PPARγ2 mRNA expression is detected with β-actin mRNA as internal control. Lane 1 displays that there is no PPARγ2 expression in the pre-differentiated hESCs. Lane 2 displays that there is PPARγ2 expression in the differentiated adipocytes.
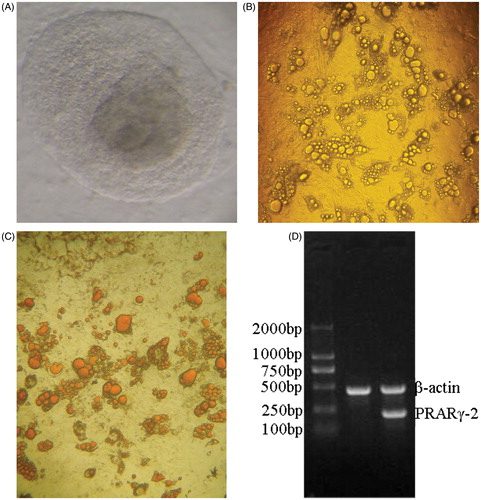
Figure 2. Pseudo-color diagrams of microarray hybridization of three polycystic ovary syndrome (PCOS)-derived adipocytes and verification of differential genes with RT-PCR. (A) cy5 is indicated with red color and cy3 with green color. For the signal in a point, if cy3 is strong, the point is indicated with green color; if cy5 is strong, the point is indicated with red color; if the intensity is similar between cy5 and cy3, the point is indicated with yellow color. Panels a, b, and c, indicate the microarray hybridization diagrams of three PCOS-derived adipocytes after comparison with non-PCOS-derived adipocytes. (B) Verification of differential genes with RT-PCR. Lane 1–4: house-keeping gene glyceraldehyde-3-phosphate dehydrogenase with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 5–8: UDP glucuronosyltransferase 2 family, polypeptide B28 gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 9–12: nuclear receptor subfamily 0, group B, member 2 gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 13–16: GALNT9(UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 9) gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 17–20: hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 21–24: tetraspanin 8 gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Lane 25–28: major histocompatibility complex, class II, DR beta 3 gene with 1st-cDNA as templates in experimental group 1, 2, and 3, and control group, respectively. Marker TaKaRa DL2000, band size are 100 bp, 250 bp, 500 bp, 750 bp, 1000 bp, 2000 bp from the top to the bottom, respectively. NR0B2: nuclear receptor subfamily 0, group B, member 2.
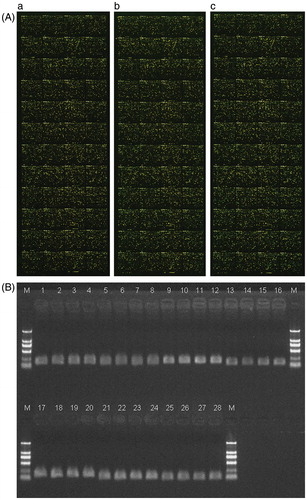
Table 1. Glycometabolism, lipid metabolism, and steroid hormone-related genes.
Table 2. Results of RT-PCR and microarray.
Table 3. Genes validated with real-time PCR and primer sequences.
