Figures & data
Figure 1. Kimura 2-parameter (K2P) distance neighbor-joining tree of 35 COI sequences from H. johnsonii (NT = New Zealand–Tasmania; Ot = other regions), five sequences from A. microlepis (Am) and 21 sequences from A. rostrata (Ar). BOLD accession numbers are given for each specimen followed by a sample ID, and location abbreviation (HM, Heard and McDonald Islands; MI, Macquarie Island; NA, North Atlantic Ocean; NP, North Pacific Ocean; NZ, New Zealand; RS, Ross Sea; SS, South Sandwich Islands); five sequences are taken from Genbank, H. johnsonii: FJ164639 and EU148182, A. rostrata: EU148703/74/75. The tree has been rooted with the morid outgroup L. microcephalus. Numbers at nodes are bootstrap percentages (>70%) after 1000 replicates, based on distance and Bayesian posterior probability values (>90%); scale bar is for K2P distance.
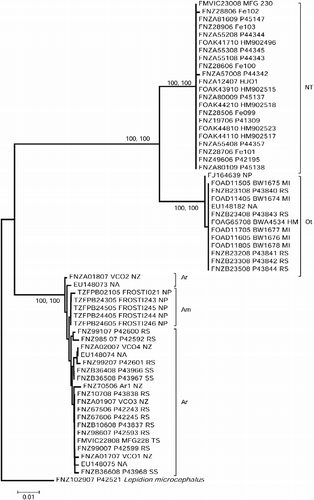
Table I. Nucleotide distance (% Kimura two-parameter) within and between species of H. johnsonii, A. rostrata, A. microlepis, and L. microcephalus.
Table II. Meristic counts from H. johnsonii from the Southern Ocean just north of the Ross Sea and New Zealand, along with values given in Gon and Heemstra (Citation1990) and Paulin (Citation1983).