Figures & data
Figure 1. Study system. Unlike most large rivers, the Congo lacks a delta, and much of the African continent's interior drains into the Atlantic through a narrow stretch of river comprising the world's greatest rapids. For 350 km, between the twin capitals of Brazzaville and Kinshasa to Boma, the Congo drops approximately 280 m in a series of 66 major cataracts. Labeo for this study were collected at localities demarcated with stars. Black, proximity of Kisangani; dark gray, Lulua River; light gray, West Africa and Lower Guinea; and white (boxed), Lower Congo rapids.
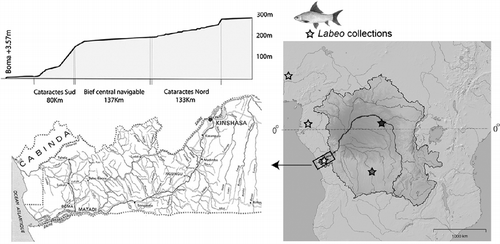
Table I. Fishes recorded from the LCR.
Table II. Specimens, sample identification codes, and collecting localities for Labeo from the LCR, Lualua River, Middle Congo (Kisangani), and Cameroon.
Figure 2. COI Labeo gene tree. Maximum-likelihood phylogram showing branch bootstrap support above the 70% (log-likelihood score of best tree: − 4137.464790). Outgroup taxa omitted. Circle, Asian Labeo; “a” and “b”, the two main clades discussed in the text. Specimen codes ending in “K” denote individuals caught in the vicinity of Kisangani (Upper Congo); “L” collected from the Lualua, a large southern tributary of the Congo River; and “C” describes specimens collected from Cameroon. One species, L. senegalensis (GenBank accession number NC008657), inhabits western Africa. The placement of L. batesii and L. annectens is in disagreement with the relationships recovered using rag1 and the control region singularly (not shown) and in concert ().
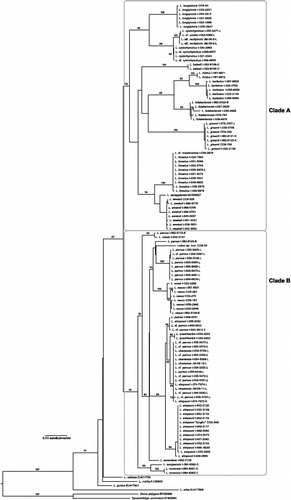
Figure 3. Phylogenetic tree of the concatenated sequence dataset (control region, COI, and rag1) of African Labeo taxa. Maximum-likelihood phylogram showing branch bootstrap support above the 70% (final GAMMA-based score of best tree: − 14613.154605). Two primary clades were recovered (A and B). The symbols denote Labeo that belong to the groupings proposed by Reid (Citation1985). Square, niloticus group; circle, macrostomus group; triangle, coubie group; diamond, forskallii group. Nine individuals represented on the COI phylogenetic tree () were omitted due to failure to amplify all genes.
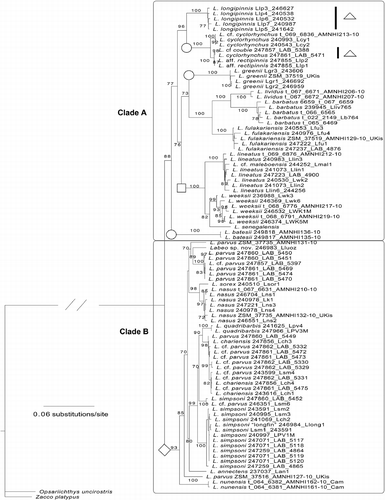
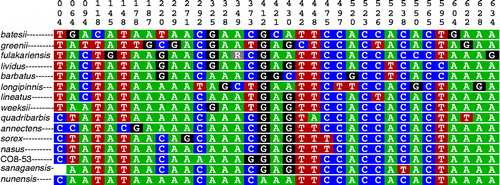
Figure 4. COI character-based key for select species of Labeo. Top vertical numbers denote positions of the 652 bp fragment analyzed. L. annectens, L. barbatus, L. batesii, L. fulakariensis, L. greenii, L. lineatus, L. longipinnis, L. nunensis, L. quadribarbis, L. sanagaensis, L. sorex, and L. sp. nov. CO8-53, with pure CAs (nucleotides that are private to that species). Labeo lividus, L. nasus, and L. weeksii with compound CAs (combinations of nucleotides).