Figures & data
Table I. Specimen data.
Figure 1. Map showing the distribution of samples of Piabina. Letters correspond to P. argentea clusters. Black square represents P. anhembi species.
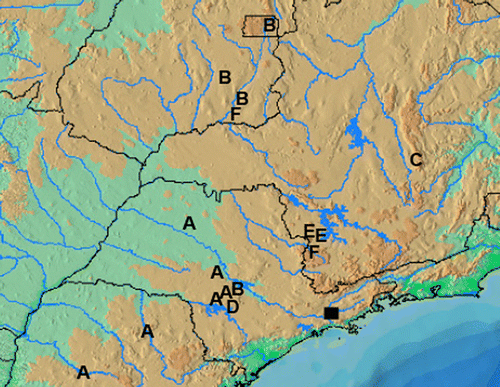
Figure 2. NJ tree of COI/CytB showing the seven major clusters obtained among Piabina specimens (A–F represent P. argentea). Node values represent statistic support: upper values, NJ bootstrap (1000 pseudo-replicates); lower values, maximum parsimony bootstrap (1000 pseudo-replicates). Numbers on fishes represent voucher number and size of photographed specimens (left and right, respectively).
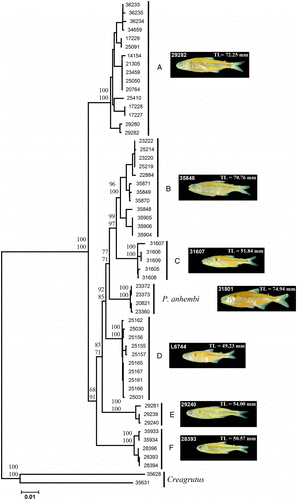
Table II. K2P genetic distance obtained among the seven major Piabina clusters.
Table III. Pairwise FST index obtained among the seven major Piabina clusters.
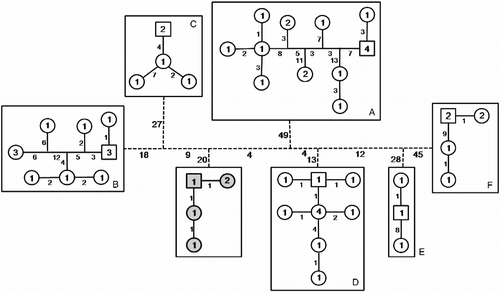
Figure 3. Seven unconnected haplotype networks among Piabina specimens. P. anhembi is represented in gray. Numbers inside the figures represent specimens that share the same haplotype. Numbers on lines represent the mutational steps between haplotypes. Dashed lines represent the necessary steps to connect the independent networks.