Figures & data
Figure 1. NJ tree of COI sequences. NJ analysis, carried out in MEGA using K2P distance, of all billfish COI barcode sequences included this study. The full tree of 353 sequences is included in Supplementary Online Material. Multiple sequences are collapsed to triangles in which vertical distance corresponds to the number of sequences, and horizontal distance is proportional to sequence diversity. Numbers indicate the number of individual specimens in a species. Several internal branches were collapsed to polytomies to indicate low bootstrap support for the same nodes in a maximum parsimony analysis—see supplementary online material. Images are taken from the FAO ‘Billfishes of the World’ catalog (CitationNakamura 1985) and are used with the permission of the copyright holder.
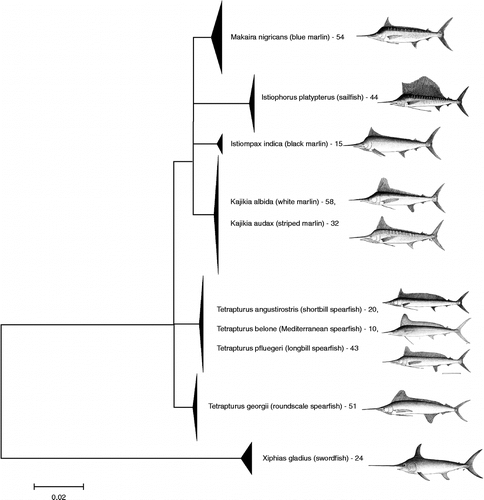
Figure 2. Haplotype parsimony network for white and striped marlin. Haplotype parsimony network, generated using the program TCS at the 99% connection limit, of 91 COI sequences belonging to the white marlin (K. albida) and striped marlin (K. audax). Individuals carrying the shared haplotype (square box) included six specimens of K. albida (EBFSF021-07, EBFSF025-07, EBFSF103-09, EBFSF115-09, MXII141-07, MXII138-07) and five of K. audax (EBFSF278-10, EBFSF280-10, EBFSF284-10, EBFSF289-10, EBFSF291-10).
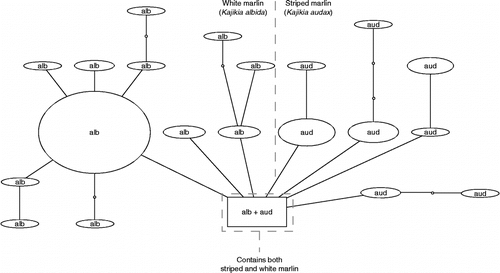
Figure 3. Haplotype parsimony network for spearfishes. Haplotype parsimony network, generated using the program TCS at the 99% connection limit, of 64 COI sequences belonging to the spearfish species T. angustirostris, T. belone, and T. pfluegeri.
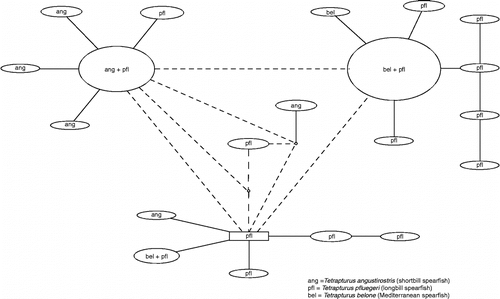
Figure 4. NJ tree of mtCR sequences. NJ analysis, carried out in MEGA using K2P distance, of mtCR sequences, extracted from GenBank and originally from the studies by Graves and McDowell (Citation2006) and McDowell and Graves (Citation2008).
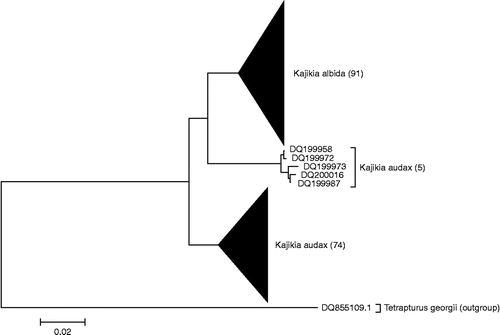
Figure 5. NJ tree of rhodopsin sequences. NJ analysis of 72 nuclear rhodopsin (Rho) sequences, carried out using the BOLD Taxon ID Tree function, using K2P distance. Arrow indicates the four apparently heterozygous K. audax individuals.

