Figures & data
Figure 1. SIRT1 protein is overexpressed in CTCL cells. (A) Immunoblot analysis for endogenous SIRT1 expression was performed using normal blood (resting PBMC), 5 CTCL cell lines (HH, Hut78, MyLa, SeAx, and SZ4), T-ALL Jurkat, and A375 melanoma (the latter from a separate blot and separated by a line). Numbers show the relative density of protein bands normalized to loading control GAPDH. Blots shown are representative of 3 independent experiments. (B) Intracellular SIRT1 was analyzed by flow cytometric analysis. Overlaid curves represent SIRT1 (black line) compared with isotype controls (filled gray). (C) Immunocytochemical staining shows SIRT1 immunoreactivity in CTCL cytopreparations, developed with DAB chromogen and methylene blue counterstain and photographed with Nikon Eclipse Ti microscope and a high-performance CCD camera using with an oil immersion lens at 60× magnification. (D) Immunohistochemical SIRT1 expression in lesional skin from CTCL patients showing clusters of stained tumor cells in epidermis and dermis (white arrow heads). Staining protocol was similar to , and images were photographed at 40× magnification using an oil immersion lens.
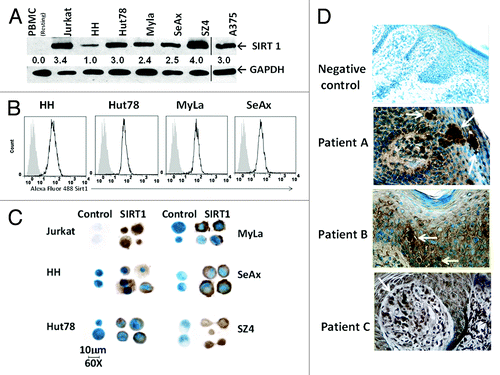
Figure 2. SIRT1 knockdown in CTCL cells results in apoptosis and decreased cell proliferation associated with FoxO3 upregulation. (A) Lentiviral shRNA-mediated knockdown of SIRT1 results in reduced SIRT1 protein expression in whole-cell lysates of CTCL cells, upregulation of FoxO3 in nuclear lysates of Jurkat and CTCL Hut78 and MyLa cells, and PARP cleavage. The vertical line shows a separate blot. For densitometric analysis, the level of endogenous SIRT1, FoxO3, and cleaved PARP levels in shNS-transduced controls was considered as 1.0. Densitometric analysis of protein bands was performed relative to GAPDH loading control. (B) shRNA-mediated knockdown of SIRT1 reduces cellular metabolic activity. White bars represent shNS-transduced samples and gray bars represent shSIRT1-transduced samples. Data represented as mean proliferation ± standard deviation of 3 experiments with similar results (*P < 0.05).
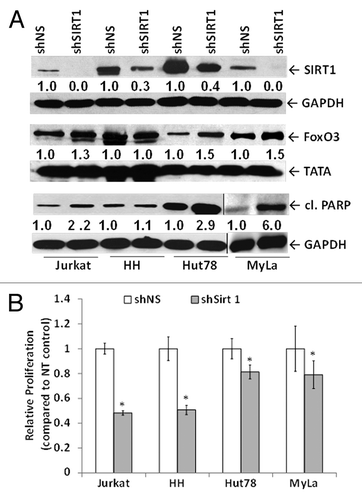
Figure 3. Chemically induced SIRT1 inhibition causes cell death in association with reduced SIRT1 enzymatic activity, increased PARP cleavage, and increased FoxO3. (A) Tenovin-1 mediated reduction in SIRT enzymatic activity. Y-axis represents percent SIRT normalized luminescent activity compared with untreated control. (B) Tenovin-1 treatment reduces the viability of CTCL cells in a dose-dependent fashion that is maximal at 24 or 48 h depending on the specific CTCL line. White bars represent untreated controls and light and dark gray bars represent tenovin-1 at 10 and 25 μM doses, respectively. Data represented as mean ± standard deviation of 3 experiments with similar results (*P < 0.05). (C) Immunoblot analysis shows tenovin-1 mediated downregulation of SIRT1 expression, at 48 h post-treatment, is associated with PARP cleavage. (D) It is also associated with increased nuclear FoxO3. Numbers represent densitometric analysis of protein bands. For each cell line, the levels of endogenous SIRT1, cleaved PARP, and FoxO3 expression in untreated controls were considered as 1.0. Densitometric analysis of protein bands was performed relative to GAPDH/TATA loading control.
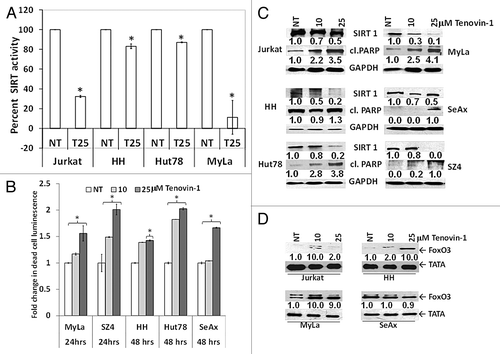
Figure 4. Tenovin-1 enhances p53 pathway signaling in wtp53 MyLa cells. (A) Immunoblot shows baseline expression levels of p53 using antibody (FL393) that detects full-length p53 of human origin. Densitometric analysis of protein bands was performed relative to GAPDH loading control. (B) Immunoblot using another antibody (7F5; 2527) specific for the N terminus of p53 shows that p53 is upregulated only in the cell line bearing wtp53 (MyLa). (C) Tenovin-1 treatment upregulates activated (acetylated) p53 expression only in wtp53 MyLa nuclear lysates. Densitometric analysis of protein bands was performed relative to TATA-binding protein (TATA) loading control. (D) Tenovin-1 treatment upregulates p53 promoter activity in wtp53 MyLa cells. Data represented as mean ± standard deviation of 3 experiments (6 replicates per experiment) with similar results (*P < 0.05). (E) Immunoblot shows that the upregulated p53 in MyLa cells is associated with upregulation of p21, a downstream target of p53. Densitometric analysis of protein bands was performed relative to GAPDH loading control.
Figure 5. Effects of combined class I/II and class III HDACIs in CTCL. (A) Combination of different classes of HDACIs results in more cell death in wtp53 MyLa than either single agent. Statistical significance (P < 0.05) when compared with no treatment control (*), 25 μM tenovin-1, (!) and 5 μm vorinostat (^). (B) Increased transcriptional activity of p53 in the wtp53 MyLa CTCL line was greatest with combination of both classes of HDACIs. The histogram represents statistical significance when compared with no treatment control (*), 25 μM tenovin-1, (!) and 5 μm vorinostat (^) in 3 experiments with similar results (P < 0.05).
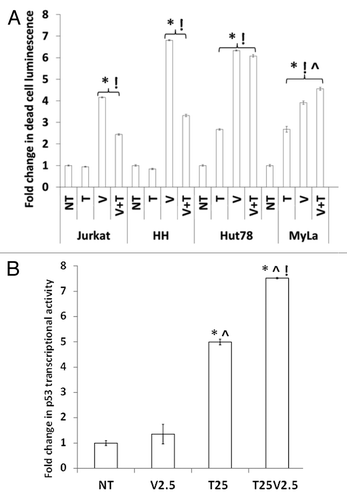
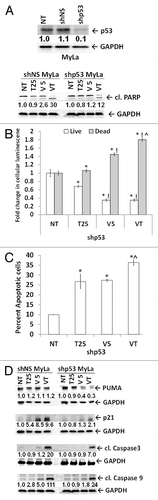
Figure 6. The p53 knockdown in MyLa cells blunts the effects of HDACIs. (A) MyLa cells (NT), with p53 knocked down by lentiviral control (shNS) and shp53. Tenovin-1, vorinostat, and combination-treated shNS and shp53 MyLa cells showed treatment-dependent apoptosis by cleaved PARP (89 kDa band). GAPDH served as loading control. (B) HDACI mediated increase in dead cell luminescence (gray bars) and decrease in live cell luminescence (white bars) compared with no treatment (NT) control (*), T25 (tenovin-1; 25 μM) (!), V5 (vorinostat; 5 μm) (^), VT (combination of V5 and T25). (C) shp53 MyLa cells treated as in 6B, stained with Annexin V and PI and analyzed by flow cytometry for apoptosis. (D) Immunoblot analysis shows the effect of single and combination treatments on protein levels of PUMA, p21, cleaved caspase-3 and cleaved caspase-9 and in shNS and shp53 MyLa cells. Densitometric analysis of protein bands was performed relative to GAPDH loading control.