Figures & data
Figure 1. IL-15 signaling in CTCL (A) Left: In normal T cells, IL-15R activates the Jak3/STAT3 pathway, which can result in specific cytokine secretion. The specific cytokine(s) expressed depends on the activation and differentiation status of the T-cell type in question. Right: In malignant T cells from Sézary syndrome patients, IL-15 activates the Jak3/STAT3 signaling pathway, which, in turn, initiates transcription of the gene encoding IL-17A. (B) In MF it is unknown whether (1) the constitutive activation of the Jak3/STAT3 is induced by IL-15; and (2) STAT3 drives the constitutive transcription of IL-15. In other words, whether IL-15 is part of an autocrine stimulation loop involving the Jak3/STAT3 pathway and downstream target genes IL-17F and IL-15.
Figure 2. IL-15 expression in malignant T cells. (A) Non-malignant (MySi) and malignant (PB2B, MF2000) T cells were cultured for 24 h. RNA was purified from the cells and reversely transcribed to cDNA and analyzed by quantitative PCR to determine the relative level of IL-17F and GAPDH mRNA. In each sample, the level of IL-17F mRNA was normalized to the amount of GAPDH mRNA and depicted as fold change when compared with non-malignant T cells (MySi). (B) Malignant (PB2B) T cells were cultured with either DMSO, Jak3 inhibitor (40 ug/mL), Jak inhibitor (1 mM), Sta-21 (40 uM), or Ag1478 (200 ng/mL). Cells were harvested after 4 h, and RNA was purified and reverse transcribed to cDNA and analyzed by quantitative PCR to determine the relative level of IL-15, IL-17F, and GAPDH mRNA. In each sample, the level of IL-15 and IL-17F mRNA was normalized to that of GAPDH mRNA and depicted as percent inhibition when compared with cells cultured with DMSO.
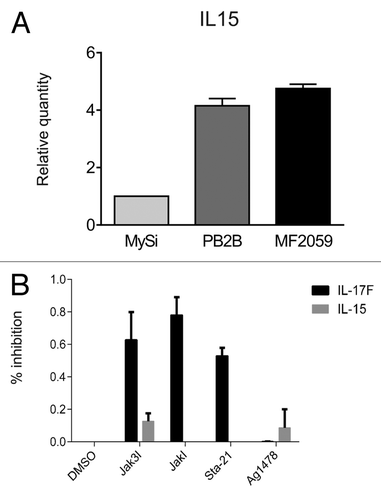
Table 1. Characteristics and phenotypes of cell lines
Figure 3. IL-15 drives STAT3 activation in non-malignant but not in malignant T cells (A) Non-malignant (MySi) and malignant (MF2000) T cells were cultured with or without IL-15 neutralizing antibody (IL-15 Ab, 2 μg/mL) for 30 min. Then, IL-15 (10 ng/mL) was added as given and the cells cultured for 30 min or 4 h further. Finally, the cells were lysed and the lysates analyzed by western blotting using antibodies against pYStat3, Stat3, and Erk1/2. (B) Malignant T cells (MF2000) were cultured with and without IL-15 Ab (2 μg/mL) for 30 min, 4 h, or 24 h. Subsequently RNA was purified from the cells and reverse transcribed to cDNA that was subjected to quantitative PCR analysis to determine the relative level of IL-17F and GAPDH mRNA. In each sample, the level of IL-17F mRNA was normalized to that of GAPDH mRNA and depicted as fold change when compared with cells cultured without IL-15Ab for the same period of time.
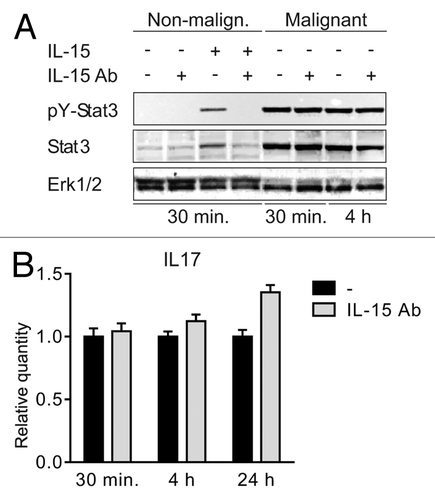
Figure 4. STAT3/IL-17F expression is driven by an IL-15-independent pathway. (A and B) Malignant (MF2059) T cells were transiently transfected with small interfering RNA against IL-15 or non-target control (NT) and cultured for 24 h. After incubation, RNA was purified from the cells and reverse transcribed to cDNA and analyzed by quantitative PCR to determine the relative level of IL15, IL17F, and GAPDH mRNA. In each sample, the level of IL15 mRNA or IL17F mRNA was normalized to the amount of GAPDH mRNA and depicted as fold change when compared NT control. There was a significant difference in IL-15 expression (P value = 0,002). (C) Malignant (MF2059) T cells were transiently transfected with small interfering RNA against IL-15 or non-target control (NT). Twenty-four hours after transfection, cells were washed and cultured for another 24 or 48 h. The cells were lysed and the lysates analyzed by western blotting using antibodies against pY-STAT3, STAT3, and actin.
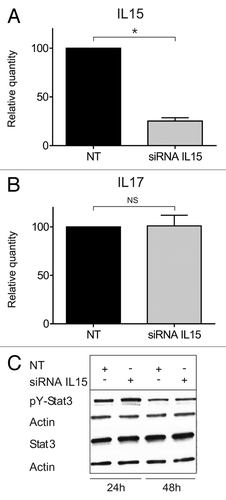
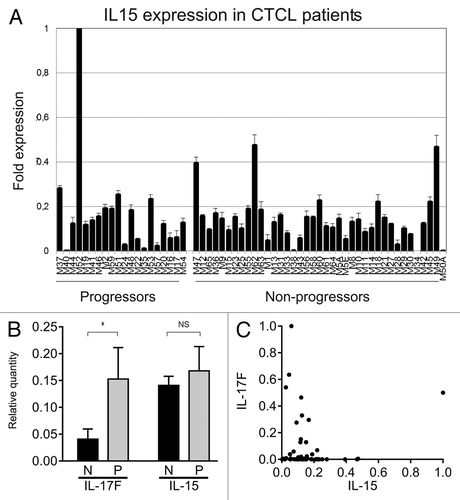
Figure 5. IL-15 mRNA in skin lesions from MF patients. (A) IL-15 mRNA expression from skin lesions in 60 MF patients. The patients, the purification of RNA from the skin biopsies, and the method of determination of the relative cytokine expression by QPCR have been described extensively previously .Citation8,Citation30,Citation37–Citation38 (B) The average relative expression of IL-17F and IL-15 mRNA in skin biopsies from CTCL patients with non-progressive (N) and progressive (P) disease. Bars represent mean + SEM of the 60 patients. *Denotes a significant difference (P < 0,05) whereas NS indicates no significant change. (C) Scatter plot of the relative mRNA expression of IL-17F and IL-15 in skin biopsies from 60 CTCL patients as above. (B and C) The depicted IL-17F expressions are based on RT-PCR data obtained in a previous study,Citation8 whereas the IL-15 expressions are novel RT-PCR data.