Figures & data
Figure 1. Mechanisms regulating the anterograde and retrograde trafficking of Panxs. (A) Anterograde trafficking of Panx1 begins in the endoplasmic reticulum (ER), where it receives a post-translational modification (PTM) in the form of a high-mannose glycan to asparagine 254 (N254) in its second extracellular loop. This forms the high mannose glycosylation species of Panx1 (Gly1). The Gly1 Panx1 species is transported to the Golgi through anterograde COPII vesicle-mediated transport. Here the high mannose N-linked glycan is modified to a complex glycosylation (Gly2). Gly2 glycosylation is necessary for subsequent plasma membrane localization of Panx1. Once at the cell surface, an interaction between the Panx1 C-terminus and filamentous actin (F-actin) is critical for stability at the plasma membrane. The intrinsic and extrinsic cues for retrograde trafficking remain poorly understood (denoted by ‘?’), however there is experimental evidence for Panx1 degradation within endolysosomal compartments. (B) The anterograde trafficking of Panx2 begins in the endoplasmic reticulum. Here, Panx2 receives a high-mannose glycan at an unknown amino acid locus. Panx2 is not glycosylated further and distributes primarily to intracellular compartments, such as the endolysosome, with evidence of cell type-specific plasma membrane localization. The mechanisms controlling anterograde and retrograde Panx2 trafficking remain unknown. (C) Anterograde trafficking of Panx3 also begins in the ER, where it is glycosylated at asparagine 71 (N71) in its first extracellular loop, forming a high mannose glycosylation species (Gly1). Like Panx1, Panx3 is trafficked to the Golgi in COPII transport vesicles, where it is modified to a mature, complex glycosylation species (Gly2). Gly2 glycosylation is necessary for trafficking of Panx3 to the plasma membrane. The internalization and retrograde transport mechanisms remain unknown, however based on endolysosomal localizations of the other Panx family members, Panx3 may also traffic in retrograde to these compartments.
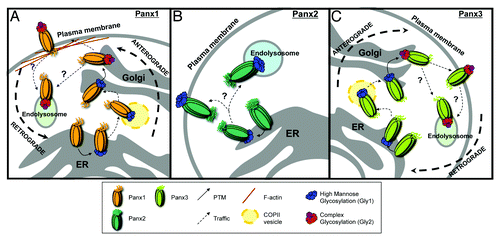
Figure 2. Panxs contain putative endocytotic recognition sequences and endolysosomal targeting sequences. Each Panx subunit contains four transmembrane domains, two extracellular loops, and three intracellular domains, including an N-terminus (NH2), intracellular loop, and a highly divergent C-terminus (COOH). The Panx2 C-terminus is the most highly divergent domain in terms of sequence homology and length, at over twice the length (361 amino acids – residues 316–677) of the Panx1 (128 amino acids - residues 298–426) and Panx3 C-termini (103aa – residues 289–392). Amino acid position of the predicted fourth transmembrane domain and the terminal amino acid of each C-terminus are indicated next to their corresponding domains. A combination of known signaling sequences and bioinformatics tools predict that each Panx protein contains both endolysosomal targeting sequences (BLUE; “*TRG_LysEnd_APsAcLL” - http://elm.eu.org/) and endocytotic recognition sequences (GREEN; “TRG_ENDOCYTOTIC_2” - http://elm.eu.org/) that putatively interact with GGA adaptor and AP protein complexes – key players in clathrin-mediated endocytosis.Citation49-Citation51
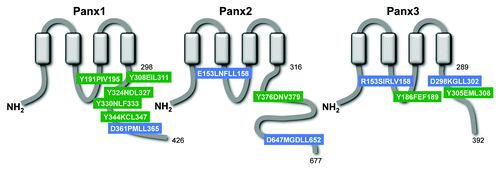
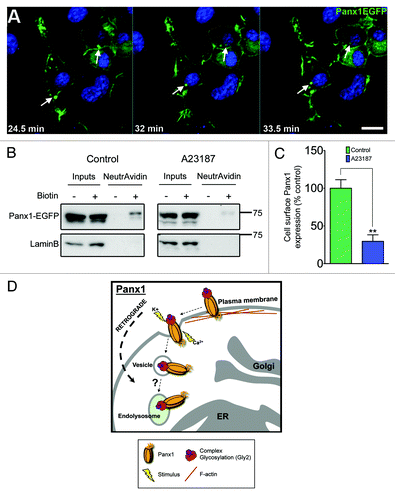
Figure 3. Panx1 activation stimulates channel internalization. (A) Selected time-lapse images from confocal live cell imaging of Panx1-EGFP expressing Neuro-2A neuroblastoma cells, demonstrates retrograde movement of EGFP-positive vesicles (arrows track same structure - images captured every minute) following extracellular elevated K+ (10mM) stimulus. Scale bar = 10μm. Hoechst 33342 was used as a nuclear stain. (B) western blotting analysis of cell surface biotinylation followed by NeutrAvidin bead pulldown illustrates Panx1-EGFP expression at the plasma membrane in Neuro-2A neuroblastoma cells. Lower levels of Panx1-EGFP were observed in the biotinylated fraction following calcimycin A23187 ionophore treatment (10µM, 4 h; right panel) compared with control (left panel). The membranes were stripped and re-probed for LaminB (nuclear) to control for any non-specific biotinylation of intracellular proteins (lower panels). These western blots are representative of three independent biological replicates. (C) Quantification of the relative levels of cell surface Panx1-EGFP under control and A23187 treatments. Calcimycin significantly reduced cell surface Panx1 levels (P = 0.0077 by unpaired Student’s t test). Western blot quantifications were performed using ImageJ. (D) We propose that upon stimulation (increased extracellular K+ or intracellular Ca2+) activated Panx1 channels are internalized and trafficked in retrograde, likely to endolysosomes.