Figures & data
Figure 1.JanusMatrix separates the amino acid sequence of T cell epitopes into TCR-facing residues and HLA binding cleft-facing residues, and then compares the TCR face to other putative T cell epitopes. JanusMatrix defines cross-reactive T cell epitopes as those that have the same MHC allele restriction, the same or similar T cell-facing residues (epitope), and conserved binding of MHC-facing residues (agretope). The HLA-facing residues of the comparators are allowed to vary, as long as they still bind to the original HLA allele. Epitopes that are identical in terms of their TCR face and are equally able to bind to the identical HLA, but differ in sequence, are rapidly identified from a given database of genomic sequences. This enables large-scale comparisons between TCR-homologous T cell epitopes from the HG, the HM, and the HP.
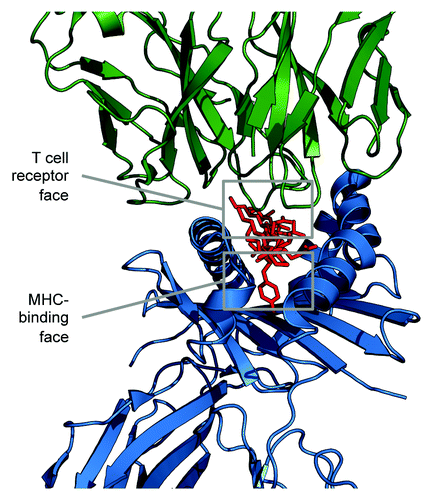
Table 1. JanusMatrix TCR-cross reactivity frequencies for three types of epitopes
Figure 2. TCR-Epitope Networks (developed using Cytoscape) for regulatory T cell epitopes (A) and effector T cell epitopes (B). Epitopes with TCR-facing residues similar to each of the test epitopes were identified in protein sequences from the human genome (HG; left), human microbiome (HM; center), and human viral and bacterial genomes (HP; right) databases. Green diamonds represent source peptides; gray squares are predicted nine-mer epitopes derived from the source peptide (predicted using EpiMatrix); blue triangles are nine-mers that are 100% identical to the TCR face of the source epitope and that are predicted to bind to the identical HLA; and light purple circles are proteins containing the cross-reactive epitope.
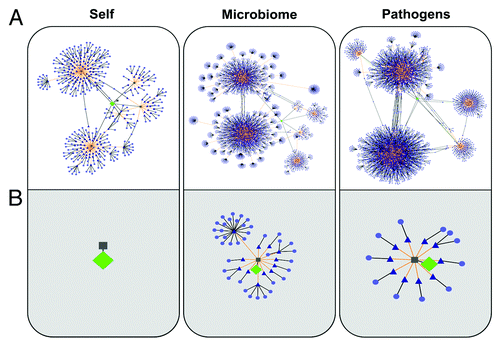
Table 2. JanusMatrix analysis of the CEFT peptide pool
Figure 3. Hand-Foot-Mouth Disease (HFMD) epitope conservation in human microbiome/human pathogen genome sequence and immunodominance. The figure shows the potential importance of cross-reactivity between microbial genomes to the immunodominance of a particular epitope. In HFMD, extensive cross-reactivity with the HM seems to predict immunodominance. Y axis: number of cross-reactive hits in the database; x axis, individual epitopes (and nine-mers within those epitopes).
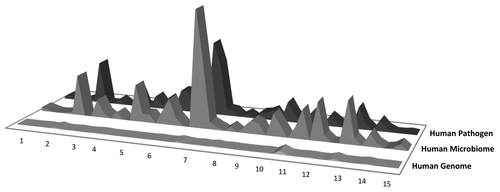
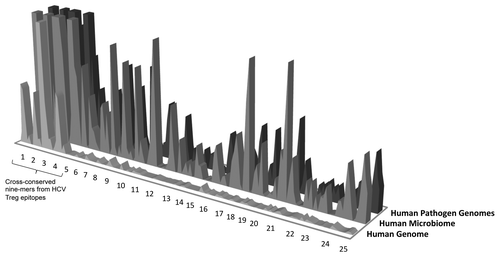
Figure 4.Hepatitis C virus (HCV) epitope conservation in human microbiome/human pathogen genome sequence and immunodominance. For regulatory T cell epitopes defined in HCV disease, HG cross-reactivity is more extensive. Overall, greater cross-reactivity with HG, HM, and HP seems to distinguish published Treg and T effector epitopes. The exact parameters defining “greater” and “lesser” cross reactivity remain to be defined following the evaluation of a number of well-defined Treg and T effector epitope examples. Y axis: number of cross-reactive hits in the database; x axis, individual epitopes (and nine-mers within those epitopes).