Figures & data
Table 1. Primers to amplify the gB, pp65, pp150 and IE1.4 genes from HCMV genome to construct individual DNA vaccines
Figure 1. Design of HCMV glycoprotein B DNA vaccine constructs with different forms of inserts. Schematic representation of the full length gB protein (amino acid 1–907) is shown at the top and DNA vaccine expressing truncated just prior the transmembrane region of gB (amino acid 1–692) is shown in the lower part of the figure (A). Expression of gB-full length and shorted gB in HEK 293 T cells (Western Blot analysis) (B). To detect of gB antigen rabbit sera collected at 1 week after forth immunization with gB-full length and gB-s were used. Cell lysates (L) and supernatants (S) from 293T cells transfected with two different gB constructs and the empty vectors (negative control) are labeled above each western blots. Molecular weight was indicated on the right. Comparison of neutralizing activity of rabbit antisera against AD169 strain of HCMV (C). Rabbits were immunized with plasmid DNA administered to individual animal at weeks 0, 2, 4, 6. Sera tested were forth bleeding from rabbit immunized with DNA vaccines: pJW4303 (negative control), pJW4303/gB and pJW4303/gB-s. 2-fold serial dilutions of antisera were incubated with 100 pfu of AD169 virus before the mixture was used to infect human primary fibroblasts. Neutralization titers shown were the highest rabbit sera dilutions at which 50% reduction of HCMV infection was achieved.
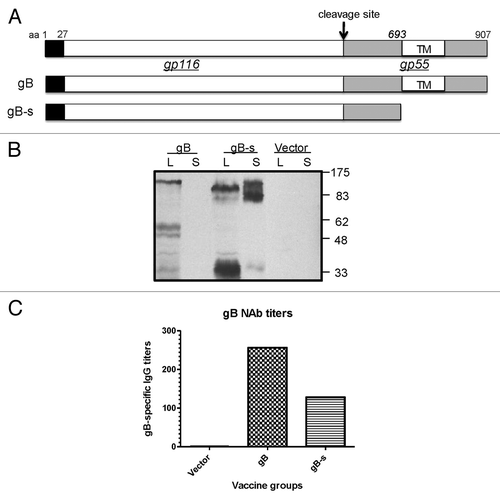
Figure 2. Schematic design of plasmid DNA constructs (A) and expression of pp65, pp150 and IE1.4 proteins in HEK 293 T cells (Western Blot analysis) (B-C) and (D) respectively). At the top we presented pp65 plasmid DNA construct (amino acid 1–561) representing full length protein; plasmid DNA construct encoding entire pp150 protein (amino acid 1–1048) was shown in the middle section and in the lower part plasmid encoding exon 4 of IE1 protein (amino acid 86–491) was presented (A). Antigen expression by pp65 and pp150 DNA vaccines was detected by pooled murine antiserum from group immunized with DNA-pp65 (B) or group immunized with DNA-pp150 vaccine (C), respectively. Expression of IE1.4 DNA vaccine construct was detected by murine monoclonal antibody, mAb p63–27 (D). Presence of pp65, pp150 and IE1.4 antigens was analyzed in supernatants (S) and lysates (L) from 293T cells. Supernatant and lysate from 293T cells transfected with vector alone was used as negative control. Molecular weight was indicated on the right.
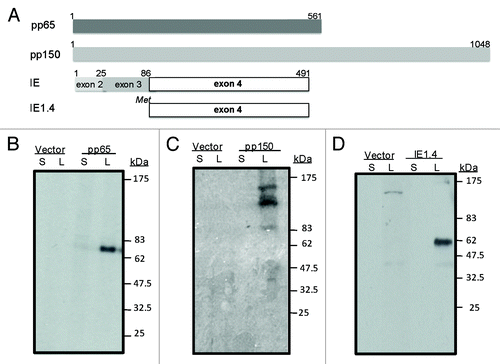
Figure 3. Production of IFN-gamma tested by ELISpot in splenocytes received from mice immunized with DNA-pp65 vaccine alone (A) or DNA-pp150 vaccine alone (B). Mice were immunized with 3x DNA vaccine encoding pp65 or pp150 antigens. Splenocytes from group of mice immunized with DNA-pp65, DNA-pp150 or vector alone were stimulated with rVV-pp65 (A) or rVV-pp150 (B). As control all immunization groups were stimulated with VV-WR.
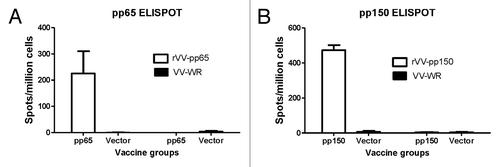
Figure 4. T cell response in mice immunized with DNA-pp65 vaccine alone tested by ICS assay. Representative dot plots show the percentage of IFN-g positive CD8+T cells (A) and IFN- g positive CD4+T positive cells (C). Lymphocytes gated on CD3+ T cells were further gated on CD8+ or CD4+ T lymphocytes. Splenocytes were stimulated with rVV-pp65 (top panel) and with VV-WR (bottom panel) (A) and (C). The levels of specific CD8+T cells are shown as percentage of IFN-gamma positive CD8+T cells (B) or IFN-gamma positive CD4+ T cells (D) in response to rVV-pp65 or VV-WR in two immunization groups (mice immunized with DNA pp65 vaccine or vector alone vaccine).
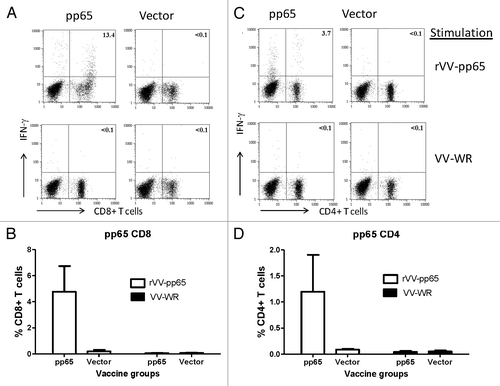
Figure 5. Induction of pp150 CD8+T lymphocytes and CD4+T cell lymphocytes following immunization with DNA pp150 vaccine alone. Representative analysis of intracellular cytokine staining assay on splenocytes stimulated with rVV-pp150 (top panel) and VV-WR (bottom panel) was shown for CD8+T cell population (A) and CD4+ T cell population (C). Numbers on the CD8+ and CD4+T–lymphocyte plots denote the percentage of the population expressing IFN-gamma (A and C). Lymphocytes gated on CD3+ T cells were further gated on CD8+ or CD4+ T lymphocytes. The levels of specific CD8+T cells in two immunization groups are demonstrated as percentage of IFN-gamma positive CD8+T cells (B) or IFN-gamma positive CD4+ T cells (D) in response to rVV-pp150 or VV-WR.
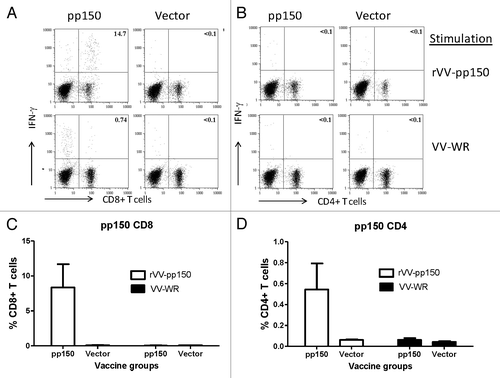
Figure 6. Immunization groups and DNA vaccines components. At week 0 and 4 mice were primed with the following vaccines: CMV alone, vector alone, DNA-1 alone, DNA-1+ CMV, DNA-2 alone or DNA-2+ CMV. At week 4 animals were boosted with either CMV or DNA-1 or DNA-2 immunizations.
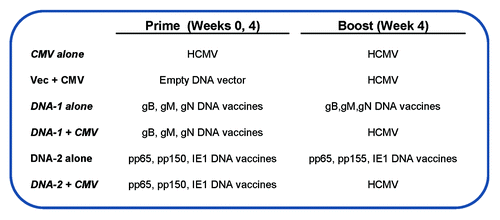
Figure 7. Detection of antibodies specific to gB (A) and gM (B) antigens by ELISA. gB-specific and gM- specific IgG titers were measured in mice sera collected after: 3 immunizations with DNA-1 alone, 2 immunizations with DNA-1 plus CMV, 3 immunizations with CMV alone or 2 immunizations with vector alone plus CMV. Data are shown as geometric mean titers within the group. P values indicate statistically significant differences measured by T test.
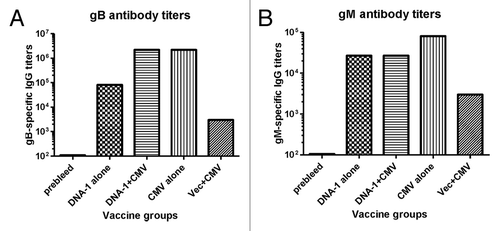
Figure 8. T cell response specific to gB antigen. Detection of gB-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of gB-SCs (B). An example of the spots generated in response to VV-gB is represented for four groups of mice immunized with: 3x DNA-1 alone (gB/gM/gN), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. As negative control splenocytes from naïve mouse were used (A). The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. Data are shown as geometric mean titers within the group. P values indicate statistically significant differences measured by T test.
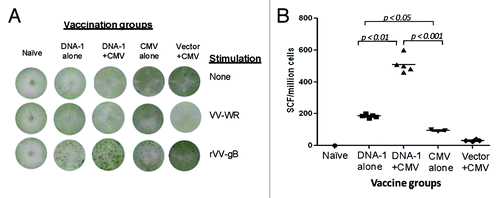
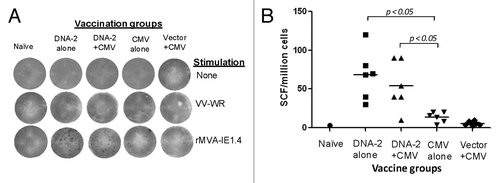
Figure 9. IFN-gamma producing T cell response specific to pp65 antigen. Detection of pp65-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of pp65-SCs (B). Representative example of spots generated to VV-pp65 in four groups of mice immunized with 3x DNA-2 alone (pp65/pp150/IE1.4), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. Splenocytes from naïve mouse were used as negative control. The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. The student T test was used to compare frequencies between groups and p values are depicted in the panel.
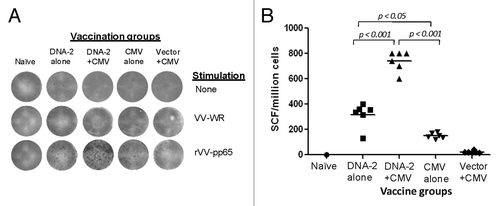
Figure 10. Cell mediated response against IE1.4 antigen measured by production of IFN-gamma in mouse splenocytes. Detection of IE1.4-SCs producing IFN-gamma measured by ELISpot assay (A) and enumeration of IE1.4-SCs (B). An example of the spots generated in response to VV-IE1.4 is represented for four groups of mice immunized with: 3x DNA-1 alone (pp65/pp150/IE1.4), 2x DNA-1 plus CMV, 3x CMV-alone and 2x vector alone plus 1x CMV. As negative control splenocytes from naïve mouse were used (A). The mean numbers of antigen-specific spot forming cells after background subtraction of control wells with no antigen were plotted (B). Experiments were conducted in triplicate. The student T test was used to compare frequencies between groups and p values are depicted in the panel.