Figures & data
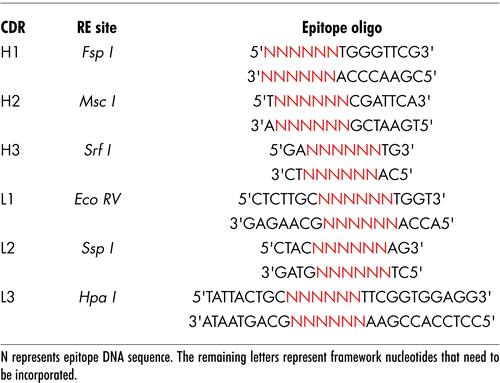
Figure 1 Construction of the ImmunoBody™ double expression vector pDCOrig. (A) Heavy chain vector pOrigHIB. The wild type DeImmunised VH region of antibody SC100 was cloned using HindIII/AfeI in frame with the human IgG1 Fc constant region. The Fc region comprises the CH1, CH2, CH3 domains and the hinge region. High-level expression in mammalian cells is driven from the CMV immediate early promoter. BGH polyadenylation signals downstream of the OrigHIB human IgG1 chain to ensure mRNA stability and effective termination. EM7 is a bacterial promoter that controls expression of the zeocin resistance gene allowing antibiotic selection in E. coli while the SV40 early promoter upstream of the resistance gene allows selection in mammalian cells. SV40 polyadenylation signals downstream of the resistance gene in order to direct proper processing of the 3′end of the zeor mRNA. The vector also contains within its backbone the ColE1 origin of replication for propagation in bacteria. (B) Light chain vector pOrigLIB. The wild type deimmunized VL region of antibody SC100 was cloned using BamHI/BsiWI in frame with the human kappa constant region. High-level expression in mammalian cells is driven from the CMV immediate early promoter. BGH polyadenylation signals downstream of the OrigLIB chain to ensure mRNA stability and effective termination. The vector also includes the ColE1 origin of replication and the antibiotic resistance gene for ampicillin allowing propagation and selection in bacteria. (C) Double expression vector pDCOrig. Once all epitopes have been incorporated into the VH and VL sites within the single vectors they are transferred into the double expression vector utilizing as highlighted HindIII/AfeI and BamHI/BsiWI in frame with their respective human constant regions. The Fc region of the heavy chain comprises of the CH1, CH2, CH3 domains and the hinge region. High-level expression of both the heavy and light chains in mammalian cells is driven from the CMV immediate early promoter. (D) Complimentary determining DNA sequences were removed by overlapping PCR and exchanged for the unique restriction sites RE1, RE2 and RE3 (EcoRV, SspI and HpaI) within the single light chain and heavy chain constructs RE4, RE5 and RE6 (FspI, MscI and SrfI respectively) singly and in combination.
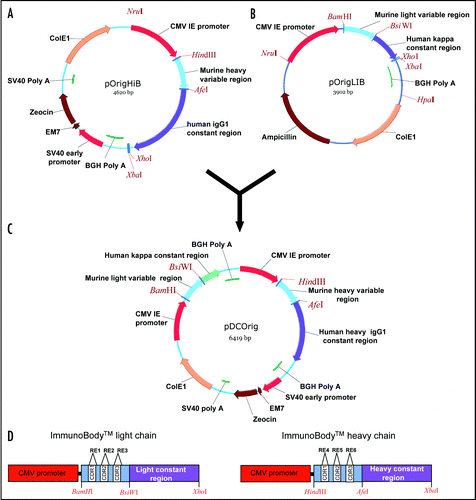
Figure 2 Antibody secretion. (A) A total of 2 µg of purified protein (lanes 1, 3 and 5) and supernatant from transfected CHO-S cells (lanes 2, 4 and 6) were loaded onto a 12% SDS-PAGE gel and subjected to electrophoresis under non-reducing conditions. Wild type ImmunoBody™ antibody is shown in lanes 1 and 2, TRP2 grafted into the CDRH2 site alongside the gp100 210M CTL in CDRH1 and HepB help CD4 epitope in CDRL1 in lanes 3 and 4 and the TRP2 CTL epitope in CDRH3 in lanes 5 and 6. The nitrocellulose blot was incubated with a HRP goat anti human IgG Fc specific antibody. (B) Western blot analysis was carried out as above however the nitrocellulose blot was incubated with a HRP anti human kappa light chain antibody. (C) Hydropathicity plots of CDRH3 within the wild type ImmunoBody™ heavy chain and those incorporating CTL (TRP2) and CD4 (HepB help) epitopes. The Kyte and DoolitleCitation70 hydropathicity index was utilized to calculate the hydropathicity distribution.
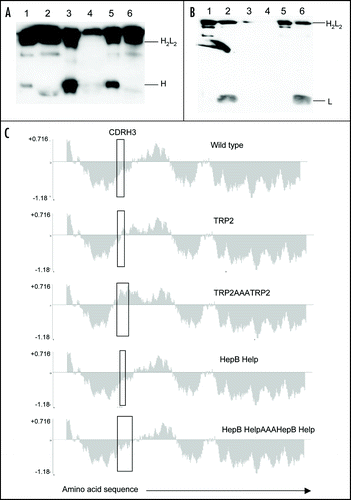
Figure 3 ImmunoBody™ DNA immunization. (A) C57Bl/6 mice were immunized at days 0, 7 and 14 with an ImmunoBody™ protein containing the H-2Kb epitope in the CDRH3 site (n = 6) or the equivalent DNA (intact) (n = 12) or ImmunoBody™ DNA containing the H-2Kb restricted TRP2 epitope in CDRH2 (non associated; n = 9). On day 19 splenocytes from immunized mice were analyzed ex vivo for the presence of epitope specific responses. Responses are measured as spots/million splenocytes. (B) C57Bl/6 mice were immunized on days 0, 7 and 14 with an ImmunoBody™ DNA construct containing TRP2 epitope in CDRH2 with or without leader sequence. On day 19 splenocytes were analyzed by IFNγ elispot assay against TRP2 peptide in triplicate. Responses are measured as spots/million splenocytes and normalised against an irrelevant peptide control (n = 4). (C) C57Bl/6 mice were immunized at days 0, 7 and 14 with an ImmunoBody™ DNA containing the H-2Kb epitope in the CDRH2 site (n = 6) or whole murine TRP2 antigen in pcDNA3 (n = 6) or pOrig vectors (n = 6). On day 19 splenocytes from immunized mice were analyzed ex vivo for the presence of epitope specific responses. Responses are measured as spots/million splenocytes. (D) C57Bl/6 mice were immunized with human IgG1 DNA containing TRP2 epitope in CDRH2 at days 0 and 7 (n = 6), or 0, 7 and 14 (n = 6). Splenocytes were analyzed on day 19 for the presence of TRP2 epitope specific responses by IFNγ elispot. Responses are measured as spots/million splenocytes. (E) C57Bl/6 mice were immunized with ImmunoBody™ DNA at days 0, 7 and 14 via gene gun (n = 6). Responses were analyzed at day 20, day 48 post primary immunization. A parallel group of mice was boosted at day 42 (n = 6). Responses were tested in ex vivo elispot assay against TRP2 peptide and an irrelevant control in triplicate. Responses are measured as spots/million splenocytes. All results are an average of two independent experiments.
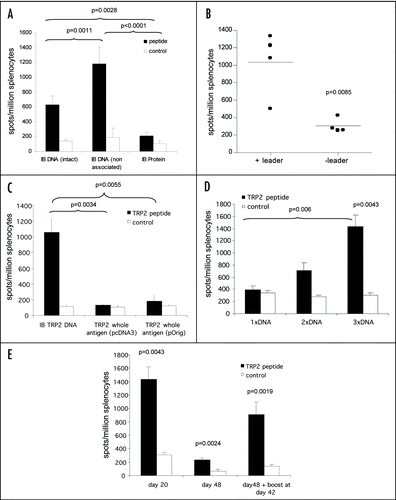
Figure 4 Linked help is required for optimal CTL responses. (A) C57Bl/6 or HLA-A*0201 transgenic mice were immunized at day 0, 7 and 14 with ImmunoBody™ heavy chain only DNA constructs containing the H-2Kb restricted TRP2 epitope in CDRH2 or the HLA-A*0201 restricted gp100 210M epitope in CDRH1. On day 19, splenocytes were analyzed by IFNγ elispot assay against gp100 210M peptide, TRP2 peptide and control. Responses are measured as spots/million splenocytes (n = 6). (B) C57Bl/6 mice were immunized at days 0, 7 and 14 with an ImmunoBody™ DNA containing the H-2Kb restricted TRP2 epitope in CDRH2, the HLA-A*0201 restricted epitope gp100 210M in CDRH1 and the I-Ab restricted HepB helper epitope in CDRL1 with either human IgG1 or murine IgG2a constant domains. On day 19, splenocytes were analyzed by IFNγ elispot assay against TRP2 peptide, HepB helper peptide and control. Responses are measured as spots/million splenocytes (n = 6). (C) HLA-DR*0401 transgenic mice were immunized at days 0, 7 and 14 with an ImmunoBody™ DNA containing the gp100 DR4 epitope in CDR H1, TRP2 epitope in CDR H2 and gp100 DR7 epitope in CDR H3 with either human IgG1 or murine IgG2a constant domains. On day 19, splenocytes were analyzed by IFNγ elispot assay against TRP2 peptide, gp100 DR4 helper peptide and control. Responses are measured as spots/million splenocytes (n = 6). (D) C57Bl/6 (n = 48), Balb/c (n = 9) or HLA-DR*0401 (n = 6) transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing either I-Ab restricted HepB, I-Ad restricted Influenza or HLA-DR*0401 restricted gp100 epitopes in CDR L1. On day 19 splenocytes were analyzed by IFNγ elispot assay against HepB, Influenza or gp100 helper peptides and an irrelevant control. Responses are measured as spots/million splenocytes. (E) C57Bl/6 (n = 6), HLA-DR*0101 (n = 6) or HLA-DR*0401 (n = 6) transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing either I-Ab restricted HepB or HLA-DR*0401 restricted gp100 or tyrosinase or HLADR* 0101 restricted triosephosphate isomerase (TPI) epitopes in CDR L3. On day 19 splenocytes were analyzed by IFNγ elispot assay against HepB, gp100, TPI or tyrosinase helper peptides and an irrelevant control. Responses are measured as spots/million splenocytes. All results are an average of at least two independent experiments.
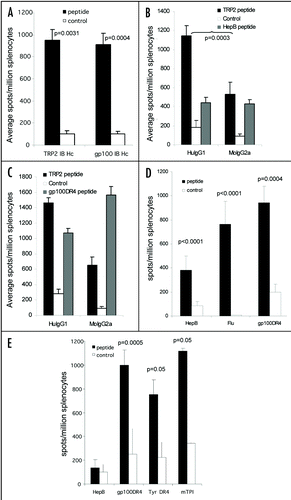
Figure 5 CTL epitopes can be processed and presented to elicit high frequency responses. (A) HLA-A2 transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing either HLA-A*0201 restricted gp100 210M (n = 12), gp100 wild type (n = 6), Mage3 (n = 6), hTERT (n = 6) or Tie2 (n = 6) epitopes in CDRH1. On day 19 splenocytes were analyzed by IFNγ elispot assay against gp100 210M, gp100 wild type, Mage3, hTERT or Tie2 peptides and an irrelevant control. Responses are measured as spots/million splenocytes. (B) C57Bl/6, Balb/c or HLA-A*0201 transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing the H-2Kb restricted TRP2 epitope (n = 50) which is also restricted through HLA-A*0201 (n = 12), the H-2Kd restricted HepB epitope (n = 6), the H-2Kb Ovalbumin epitope (n = 6) or the HLA-A*0201 restricted hTERT epitope (n = 6) in CDRH2. On day 19 splenocytes were assayed ex vivo in IFNγ elispot assay against relevant and control peptides. Responses are measured as spots/million splenocytes. (C) C57Bl/6 or HLA-A*0201 transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing the H-2Kb restricted TRP2 epitope (n = 12) or HLA-A*0201 restricted VEGFR2 epitopes (n = 6) in CDRH3. On day 19 splenocytes were assayed for presence of specific responses by IFNγ elispot assay. Responses are measured as spots/million splenocytes. (D) Splenocytes from immunized mice were depleted of CD8 T cells and analyzed against TRP2 peptide, HepB helper peptide and a media control in triplicate for the presence epitope specific responses in IFNγ elispot assay in triplicate. Responses are measured as spots/million splenocytes (n = 4). (E) HLA-A*0201 transgenic mice were immunized on days 0, 7 and 14 with an ImmunoBody™ construct containing HLA-A*0201 restricted TRP2 epitope in CDRH2, the HLA-A*0201 restricted gp100 210M epitope in CDRH1 and the I-Ab restricted HepB helper epitope in CDRL1. On day 19 splenocytes were analyzed by IFNγ elispot assay against relevant peptides and an irrelevant control in triplicate. Responses are measured as spots/million splenocytes (n = 12). All results are an average of at least two independent experiments.
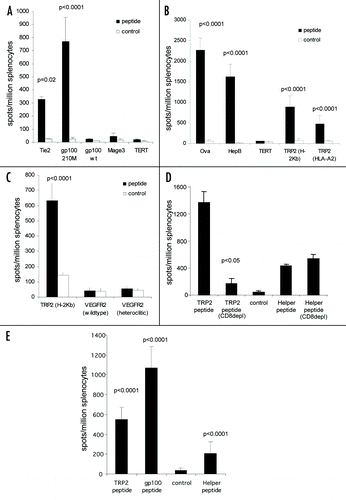
Figure 6 Immune responses generated by ImmunoBody™ DNA vaccination can delay tumour growth. HLA-DR*0401 transgenic mice were immunised with ImmunoBody™ DNA containing the H-2Kb restricted TRP2 epitope in CDRH2 and the HLA-DR*0401 restricted gp100 epitope in CDRH3 via gene gun at days 0, 7 and 14. On day 14, mice were injected s.c. with 2.5 × 104 B16F1 tumour cells. Tumour growth was monitored at 3–4 day intervals using a calliper and displayed as tumour volume at day 22 post tumour implant (n = 10).
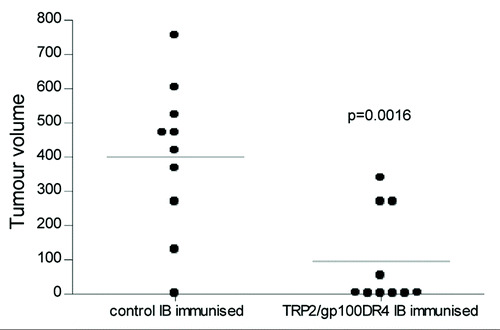
Table 1 Primers used in this study
Table 3 CTL and helper epitopes
Table 4 ImmunoBody™ protein secretion