Figures & data
Figure 1. PCNA marks DNA replication and repair sites. (A) Crystal structure of PCNA (green) encircling the DNA (red) (PDB 1AXC). PCNA serves as a loading platform for several repair and replication factors that bind to a common PCNA region indicated with a dotted line. (B) During the DNA synthesis phase (S-phase) PCNA is recruited to sub-nuclear sites of DNA replication forming distinctive patterns over time that characterize different S-phase stages (shown schematically and with confocal images of GFP-PCNA expressing cells). (C) PCNA associates with sites of DNA damage induced by laser microirradiation. Scale bar 5 μm.
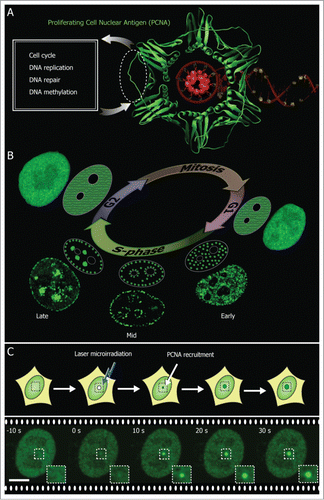
Figure 2. Design of PCNA interacting peptides (PIP). The red box highlights the residues binding to the PCNA binding pocket. The dotted lines indicate the consensus mutations introduced on the p21141–157 peptide to optimize the PIP-box sequence and obtain the p21–08 peptide. The IFS (p21158–160) sequence from p21 was removed to reduce the binding between the p21–08 and the CDK-cyclin and make its binding more selective toward PCNA. Additionally a previously describedCitation19 synthetic peptide PL derived from Pogo and DNA ligase I was used.
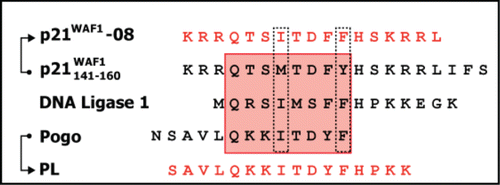
Figure 3. PCNA interacting peptides bind to PCNA in vivo but are not cell permeable. (A) Crystal structure of the PCNA trimer (green) with the C-terminus region of the p21 protein (red) bound to it (PDB ID 1AXC). The sequences of the p21 peptide and the two highest PCNA binding affinity peptides p21–08 and PL screened are displayed. (B) Structures of the p21 peptides and the two PIPs, i.e., p21–08 and PL sequences obtained from molecular dynamics simulations are shown. (C) Images showing that these peptides ([F-p21], [F-p21–08], [F-PL]) after microinjection clearly co-localize with GFP-PCNA at replication sites. (D) When the peptides are added to the extracellular media they are not able to enter the cells. The abbreviation PC stands for phase contrast and DIC for differential interference contrast. Scale bar 5 μm.
![Figure 3. PCNA interacting peptides bind to PCNA in vivo but are not cell permeable. (A) Crystal structure of the PCNA trimer (green) with the C-terminus region of the p21 protein (red) bound to it (PDB ID 1AXC). The sequences of the p21 peptide and the two highest PCNA binding affinity peptides p21–08 and PL screened are displayed. (B) Structures of the p21 peptides and the two PIPs, i.e., p21–08 and PL sequences obtained from molecular dynamics simulations are shown. (C) Images showing that these peptides ([F-p21], [F-p21–08], [F-PL]) after microinjection clearly co-localize with GFP-PCNA at replication sites. (D) When the peptides are added to the extracellular media they are not able to enter the cells. The abbreviation PC stands for phase contrast and DIC for differential interference contrast. Scale bar 5 μm.](/cms/asset/9858aa8b-b3c8-405a-b46e-a4a86767d2a8/kncl_a_980688_f0003_c.gif)
Figure 4. Rationale to develop a cell permeable DNA replication and repair marker by targeting PCNA. (A) The nucleus of a cell expressing fluorescently labeled PCNA was microirradiated. PCNA is recruited to damaged DNA sites. Fusion of the PIP with a CPP [F -TAT-p21–08] makes it cell permeable. However, the CPP sequence interferes with the PIP binding to PCNA. (B) Design strategy: the PIP is coupled to a cell-penetrating peptide (in this case the TAT peptide) via a disulfide bridge; after transporting the PIP into the cell the disulfide bridge is reduced; the CPP accumulates mainly in the cytosol and nucleolus, while the PIP is free to reach and bind PCNA. (C) To visualize in live cells the separation of the PIP from CPP, we labeled each sequence with a different fluorescent dye. The TAT peptide labeled with TAMRA is coupled by a disulfide bridge to the PIP peptide labeled with FITC [R-TAT(-SS-)p21–08-F]. It can be seen that the free PIP labels the site of DNA damage, whereas the CPP distributes mainly in the cytosol and nucleolus. Experiments were repeated at least two times. Scale bar 5 μm.
![Figure 4. Rationale to develop a cell permeable DNA replication and repair marker by targeting PCNA. (A) The nucleus of a cell expressing fluorescently labeled PCNA was microirradiated. PCNA is recruited to damaged DNA sites. Fusion of the PIP with a CPP [F -TAT-p21–08] makes it cell permeable. However, the CPP sequence interferes with the PIP binding to PCNA. (B) Design strategy: the PIP is coupled to a cell-penetrating peptide (in this case the TAT peptide) via a disulfide bridge; after transporting the PIP into the cell the disulfide bridge is reduced; the CPP accumulates mainly in the cytosol and nucleolus, while the PIP is free to reach and bind PCNA. (C) To visualize in live cells the separation of the PIP from CPP, we labeled each sequence with a different fluorescent dye. The TAT peptide labeled with TAMRA is coupled by a disulfide bridge to the PIP peptide labeled with FITC [R-TAT(-SS-)p21–08-F]. It can be seen that the free PIP labels the site of DNA damage, whereas the CPP distributes mainly in the cytosol and nucleolus. Experiments were repeated at least two times. Scale bar 5 μm.](/cms/asset/8aef6d58-d711-44cf-be78-3960b55724e0/kncl_a_980688_f0004_c.gif)
Figure 5. In vivo immediate labeling of DNA repair sites. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells were microirradiated. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide [TAT(-SS-)PL-R] was added to the extracellular media. PIP labels PCNA at repair sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 5. In vivo immediate labeling of DNA repair sites. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells were microirradiated. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide [TAT(-SS-)PL-R] was added to the extracellular media. PIP labels PCNA at repair sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/7041264a-568c-4e0b-a0f1-500d37a30e81/kncl_a_980688_f0005_c.gif)
Figure 6. In vivo immediate labeling of DNA replication sites in S-phase. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells at different cell cycle stages recognizable by the PCNA sub-nuclear patterns were identified and the same peptide as in , PL coupled to TAT peptide via disulfide bridge [TAT(-SS-)PL-R], when added to the extracellular media, labeled the sites of active DNA replication. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 6. In vivo immediate labeling of DNA replication sites in S-phase. Cartoon representation of the experimental protocol and time-lapse snapshots obtained using confocal microscopy are illustrated. GFP tagged PCNA expressing cells at different cell cycle stages recognizable by the PCNA sub-nuclear patterns were identified and the same peptide as in Figure 5, PL coupled to TAT peptide via disulfide bridge [TAT(-SS-)PL-R], when added to the extracellular media, labeled the sites of active DNA replication. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/8308c3d7-bb49-4765-8207-f03cfd277c8c/kncl_a_980688_f0006_c.gif)
Figure 7. PCNA interacting peptide (PIP) labels replication and repair in fixed cells. (A) Here replication and repair sites can be directly visualized in fixed cells without the need of antibodies by PL peptide [F-PL] in the presence of ectopically expressed PCNA. (B) The same PL peptide clearly marks the endogenous PCNA. Experiments were repeated at least three times. Scale bar 5 μm.
![Figure 7. PCNA interacting peptide (PIP) labels replication and repair in fixed cells. (A) Here replication and repair sites can be directly visualized in fixed cells without the need of antibodies by PL peptide [F-PL] in the presence of ectopically expressed PCNA. (B) The same PL peptide clearly marks the endogenous PCNA. Experiments were repeated at least three times. Scale bar 5 μm.](/cms/asset/44b7ac40-a81b-4615-a048-64e0ad7ecece/kncl_a_980688_f0007_c.gif)
Figure 8. The cell permeable and intracellular cleavable PIP directly labels DNA replication and repair in different cell types and species. The PL peptide fluorescently labeled and coupled by a disulfide bridge to the TAT peptide was added to the extracellular media on a variety of cells as indicated, with and without laser microirradiation. In the absence of labeled PCNA the PIP allows a clear classification of S-phase replication stages and damaged DNA sites. The abbreviation DIC stands for differential interference contrast. Experiments were repeated at least three times. Scale bar 5 μm.
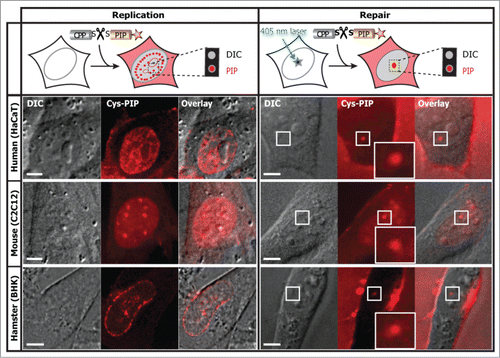
Table 1. Summary of peptides
