Abstract
Cynara cardunculus L. is a typical Mediterranean species comprising two important cultivated types, the globe artichoke (Cynara cardunculus var. scolymus) which is grown for its edible heads and the cultivated or leafy cardoon (C. cardunculus var. altilis) appreciated for its fleshy stems and leaf stalks. It includes also a third form, the wild cardoon (C. cardunculus var. sylvestris) which is considered the ancestor of the cultivated forms. Despite progress in the evolutionary field, advanced chromosome studies on C. cardunculus are almost nonexistent. The objective of this study was to fill in this gap by providing a refined cytogenetic characterization of the cultivated scolymus and altilis varieties. The karyomorphological analysis showed that artichoke and cardoon share an identical karyotype. All chromosomes are metacentric but are markedly differentiated with respect to their length, therefore they could be separated into three groups of different size: large (L), medium (M) and small (S). As a first step towards the physical mapping of artichoke and cardoon chromosomes, the FISH technique was applied to localize the position of 18S-5.8S-25S rRNA genes (45S rDNA). The fluorescent signals obtained by the FISH experiments constituted reliable landmarks for the identification of two pairs of M chromosomes and two pairs of S chromosomes. The overall results represent a significant advance in C. cardunculus cytogenetics and suggest further investigation of the wild sylvestris variety in order to acquire more exhaustive information on the evolutionary pathway of the species.
Introduction
Cynara is a relatively small genus of the Asteraceae family, native to the Mediterranean region. According to the taxonomic revision of Wiklund (Citation1992) it comprises C. cardunculus L., C. syriaca Boiss., C. auranitica Post, C. cornigera Lindley, C. algarbiensis Cosson, C. baetica Pau, C. cyrenaica Maire and Weiller, and C. humilis L., whereas Rottenberg and Zohary (Citation1996) recognize only seven species, excluding C. auranitica. The species are characterized by the same chromosome number, 2n = 34. The most important species is C. cardunculus which includes two cultivated forms, the globe artichoke (var. scolymus (L.) Fiori) and the leafy cardoon (var. altilis DC.). Only artichoke is grown on a wide commercial scale, whereas the leafy cardoon is appreciated locally in many Mediterranean countries for its fleshy stems and leaf stalks (Dellacecca Citation1990). Globe artichoke represents a typical crop of the Mediterranean area, where it contributes to the agricultural economy; however its cultivation extends also to North Africa, the Middle East, South America, USA and China. The edible parts of the plant are represented by the immature flower heads, which are consumed cooked, fresh or canned. The nutritive and therapeutic properties of artichoke have been known since ancient times; the species is rich in vitamins A, B1, B2, C and minerals, the roots contain inulin (Brown and Rice-Evans Citation1998) and the leaves are a source of antioxidants such as luteolin and di-caffeoylquinic acids (Gebhardt Citation1997). C. cardunculus comprises also a third form, the wild cardoon (var. sylvestris Lamk). Alphonse de Candolle (1886) was one of the first to perceive a close relationship among the three varieties and, from the morphological analysis of the wild Cynara species known at his time, supposed C. cardunculus var. sylvestris to be the progenitor of the cultivated types. Successively other botanists accepted this hypothesis but the definitive identification of the wild ancestor of artichoke was obtained by crossing experiments which revealed that var. sylvestris was the only wild cardoon fully cross-compatible with the cultivated variety and able to produce seeds and viable hybrids (Rottenberg and Zohary Citation1996). The evolutionary relationships between the cultivated forms and the wild cardoon have been confirmed by isozyme analysis and molecular markers such as RAPDS, AFLPs, and microsatellites and the polymorphism analysis of internal (ITS) and external (ETS) rDNA spacers (Rottenberg et al. Citation1996; Sonnante et al. Citation2002, Citation2004, 2008; Sonnante, Carluccio, et al. Citation2007). Such studies contributed to an understanding of the evolutionary pathway of the genus and the main phases of the domestication process. C. cardunculus is the most recent species and, in contrast to the other species, it is found all throughout the basin where it exhibits an extraordinary genetic variability. This aspect, along with its high adaptive capacity, facilitated the domestication process of this species (Sonnante, Pignone, et al. Citation2007). The domestication of artichoke and cardoon seems to be the result of two distinctive events. Most probably the cultivation of artichoke started at the beginning of the first millennium in Sicily, whereas that of cardoon much later, in the first half of the second millennium, probably in Spain and France (Sonnante et al. Citation2012). This evolutionary scenario is supported by molecular analyses that indicate close relationships between the artichoke and the wild cardoon from the Eastern Mediterranean, whereas cultivated cardoons are similar to the Western wild cardoons (Sonnante, Carluccio, et al. Citation2007; Sonnante et al. Citation2008). Despite such progress in the evolutionary field, the cytogenetic knowledge of C. cardunculus is still scarce, being limited to the chromosome number and little basic information on the chromosome morphology of artichoke (Giorgi et al. Citation2013; Khaldi et al. Citation2014). Molecular cytogenetic techniques, such as FISH (fluorescent in situ hybridization), which are known to be extremely valuable tools for investigating species with small chromosomes, have not been so far applied in C. cardunculus.
Therefore, this study was carried out with the intention of gaining knowledge on the chromosome background of C. cardunculus by attempting a refined cytogenetic characterization of the cultivated scolymus and altilis varieties. To this purpose a karyomorphological analysis based on chromosome measurements was carried out. To acquire further data on the chromosome structure of artichoke and cardoon and to obtain useful landmarks for chromosome identification, the FISH technique was applied to determine the number and the position of the loci of 18S-5.8S-25S ribosomal RNA genes (45S rDNA). In plants, the 18S-5.8S-25S rRNA genes are organized in many hundreds of repeated units arranged in tandem arrays in the nucleolar organizer regions (NORs) which are recognized as secondary constrictions in satellited (SAT) chromosomes. However, in species with small chromosomes, like those of C. cardunculus, such constrictions are often undetectable. The FISH technique offers the means for overcoming this limitation. The sites of 45S rDNA sequences, detected with FISH experiments during this study, constitute the first step towards the physical mapping of C. cardunculus chromosomes.
Materials and methods
Plant materials
Seeds of commercial cultivars Violetto di Sicilia and Romanesco of artichoke and Gigante di Romagna of leafy cardoon were used as experimental materials. The seeds were germinated on moist filter paper in Petri dishes and then transplanted in pots. Plants were grown in the greenhouse under appropriate temperature conditions (20–22°C) to allow a continuous production of root tips. About 15 plants for each C. cardunculus varieties have been used for this study.
Chromosome preparations and karyomorphological analysis
Mitotic chromosome preparations were performed with meristematic tissues of actively growing root tips of 5–10 mm in length. They were harvested from the potted plants and treated as described by Falistocco and Marconi (Citation2013) with only few modifications. Root tips were immersed in ice-cold water for about 24 h. After this time they were pre-treated in an 1‰ aqueous solution of a stock solution consisting of 1 ml of α-bromonaphthalene dissolved in 100 ml of absolute ethanol for 2 h and then fixed in ethanol acetic acid (3:1) overnight. Chromosome preparations for karyotype analysis and in situ hybridization experiments were performed as follows. The fixed root tips were transferred to enzyme buffer (10 mM citric acid/sodium citrate, pH 4.6) for 20 min, then they were excised and digested in the enzyme solution (4% cellulase Onozuka R10 and 1% pectolyase Sigma in distilled water) for 1 h at 37°C. Slides were prepared according to the dropping method described by Leitch et al. (Citation1994). For the karyological analyses slides stained with DAPI were used. Chromosome measurements were taken on enlarged prints of several selected metaphases. The centromeric position was calculated as the long:short arm ratio to classify chromosomes according to the system of Levan et al. (Citation1964). Karyotypes were arranged by ordering the chromosome pairs from the largest to the smallest.
Fluorescent in situ hybridization (FISH)
To establish the number and position of the 45S rDNA loci in the chromosome complements of the scolymus and altilis varieties, the clone pTa71 containing the 18S-5.8S-25S rRNA genes and non-transcribed spacers of Triticum aestivum L. (Gerlach and Bedbrook Citation1979) was used. The probe was labelled by nick translation with digoxigenin-11-dUTP. The in situ hybridization procedure was performed according to the protocol described in detail by Falistocco (Citation2009). Slides were pretreated with 100 μl ml−1 of RNase A in (0.3 M NaCl, 0.003M sodium citrate) 2× SSC and then incubated in 40 units/ml of Pepsin (Sigma, St. Louis, MO, USA) for 15 min at 37°C. The hybridization mixture, consisting of 2 ng μl−1 of the ribosomal probe, 50% (v/v) formamide, 10% (w/v) dextran sulphate, 0.1% (w/v) SDS (sodium dodecyl sulphate) and 300 ng μl−1 sheared salmon sperm DNA was incubated for 10 min at 70°C and chilled on ice. The hybridization mixture and the chromosomes were denatured together at 70°C for 5 min and allowed to hybridize overnight at 37°C. After hybridization the slides were washed in two changes of 20% formamide (v/v) in 0.1× SSC, for 5 min each. Detection of the digoxigenin labelled probe was carried out with anti-digoxigenin conjugated with FITC (Roche, Indianapolis, IN, USA). The slides were counterstained with 2 μg ml−1 of DAPI (4′,6-diamidino-2-phenylindole) and then mounted in antifade solution Vectashield (Vector Laboratories, Peterborough, UK). The slides were examined with a Microphot Nikon epifluorescence microscope (Tokyo, Japan). Photographs were taken on Fujichrome (Tokyo, Japan) 400 colour slide film and digitized with a film scanner (Nikon, Tokyo, Japan). Superimposition of images was performed with the software Adobe Photoshop. The images were treated for colour contrast and brightness uniformity when necessary.
Results and discussion
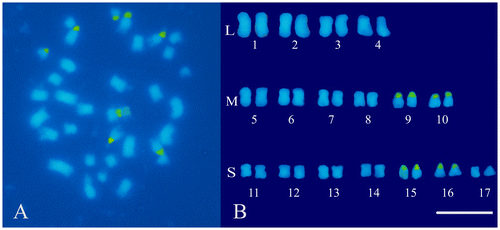
In order to achieve an effective karyomorphological analysis, initial efforts were made to obtain well-spread metaphases with an appropriate degree of chromosome contraction. For this purpose, the procedure previously used in hazelnut (Falistocco and Marconi Citation2013) was applied by introducing only a few changes in the duration of pre-treatments of the root-tips. The method used turned out to be advantageous in C. cardunculus in terms of the number and quality of the metaphases. Observations of DAPI stained preparations confirmed the chromosome number 2n = 34 for both the scolymus and altilis varieties (Figures A and A). Chromosome measurements showed that the artichoke karyotype is rather uniform with respect to the centromeric position. In fact, all chromosomes were metacentric with an arm ratio from 1.0 to 1.5. Conversely, chromosomes appeared markedly differentiated with respect to their length, which ranged from 3.2 to 1.0 μm. The total length of the haploid set was 33.8 μm. The chromosome size does not gradually decrease from the larger to the smaller chromosome pairs but discriminates three chromosome types of different size that have been designated as large (L), medium (M) and small (S) chromosomes. The mean length values of each type were 3.0, 2.3 and 1.2 μm, respectively. The karyotype was arranged by positioning the chromosome pairs into separate groups, according to their size (Figure B). Within each group variations of the chromosome size were reduced and some chromosomes appeared identical. In this case, the pairing of the chromosomes and their location within each group was arbitrary. After FISH experiments however, reliable landmarks consisting of the sites of the 45S rDNA sequences were obtained. The hybridization of the pTa71 probe generated eight distinct fluorescent signals which were localized in two pairs of M chromosomes (9 and 10) and two pairs of S chromosomes (15 and 16) (Figure B). DAPI stained metaphases of var. altilis displayed a chromosome situation similar to that discovered in var. scolymus. Also in leafy cardoon the chromosomes are metacentric (with an arm ratio from 1.2 to 1.5) and markedly differentiated in their size (Figure B). Measurements confirmed the visual analysis revealing a chromosome length variation from 3.3 to 1.0 μm. Four large chromosome pairs with a mean length of 3.0 μm were constantly distinguished from the others, while a group of six median chromosomes (2.2 μm) was recognizable from the remaining seven pairs that were notably shorter (1.2 μm). The length of the haploid complement was 34.0. For the karyotype construction, the chromosomes were arranged following the same criterion used for artichoke (Figure B). Similarly to artichoke, the 45S rDNA sequences in cardoon are distributed in four pairs of loci. The green fluorescent signals obtained by the FISH treatment were positioned at the extremity of the short arms of two pairs of median (9 and 10) and small (15 and 16) chromosomes (Figure B).
Figure 1. Somatic chromosomes of globe artichoke (C. cardunculus var. scolymus). (A) Mitotic metaphase after the FISH treatment and DAPI staining, loci of 45S rDNA are mapped in green. (B) Karyotype with chromosome pairs arranged in groups according to their size: large (L), medium (M) and small (S). Scale bar 5 μm.
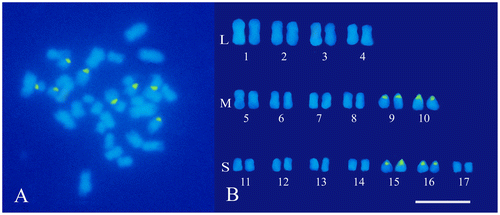
Figure 2. Somatic chromosomes of cultivated cardoon (C. cardunculus var. altilis). (A) Mitotic metaphase after the FISH treatment and DAPI staining, loci of 45S rDNA are mapped in green. (B) Karyotype with chromosome pairs arranged in groups according to their size: large (L), medium (M) and small (S). Scale bar 5 μm.
Despite the difficulty due to the small size of the chromosomes, the present study has contributed to the cytogenetic knowledge of the cultivated C. cardunculus forms. The karyomorphological analysis added new data on the chromosome structure of the artichoke to the information published by Giorgi et al. (Citation2013); moreover, information on cytogenetics of leafy cardoon has been provided for the first time. By the application of FISH, the first step towards the construction of a detailed FISH-based karyotype for C. cardunculus has been carried out. The sites of 18S-5.8S-25S rRNA genes constitute reliable cytogenetic landmarks for the identification of four pairs of chromosomes. The identification of the remaining chromosomes will be carried out by continuing the study with the use of more chromosome-specific markers. For a more comprehensive view of the evolutionary pathway of C. cardunculus, the analysis will be extended to the wild cardoon to verify the level of cytogenetic affinity between the cultivated forms and var. sylvestris.
Disclosure statement
No potential conflict of interest was reported by the author.
References
- Brown JE, Rice-Evans CA. 1998. Luteolin rich artichoke extract protects low density lipoprotein from oxidation in vitro. Free Radical Res. 29(3):247–255.
- de Candolle A. 1886. Origin of cultivated plants. 2nd ed., 1964. New York: Hafner.
- Dellacecca V. 1990. Cardo (Cynara cardunculus L.). In: Bianco VV, Pimpini F, editors. Orticoltura. Bologna: Patron; p. 252–258.
- Falistocco E. 2009. Presence of triploid cytotypes in the common fig (Ficus carica L.). Genome. 52(1):919–925.
- Falistocco E, Marconi G. 2013. Cytogenetic characterization by in situ hybridization techniques and molecular analysis of 5S rRNA genes of the European hazelnut (Corylus avellana). Genome. 56(3):155–159.
- Gebhardt R. 1997. Antioxidative and protective properties of extracts from leaves of artichoke (Cynara scolimus L.) against hydroperoxide induced oxidative stress in cultured rat hepatocytes. Toxicol Appl Pharmacol. 144(2):279–286.
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucl Acid Res. 7(7):1869–1885.
- Giorgi D, Pandozy G, Farina A, Grosso V, Lucretti S, Crinò P, Saccardo F. 2013. Karyotype of globe artichoke (Cynara cardunculus var. scolymus): preliminary studies to define its chromosome morphology. Acta Horticult. 983:33–138.
- Khaldi S, Hidalgo O, Garnatje T, El Gazzah M. 2014. Karyological and genome size insights into cardoon (Cynara cardunculus L., Asteraceae) in Tunisia. Caryologia. 67(1):57–62.
- Leitch AR, Schwarzacher T, Jackson D, Leitch IJ. 1994. In situ hybridization: a practical guide. Oxford: BIOS Scientific.
- Levan A, Fredga K, Sandberg AA. 1964. Nomenclature for centromeric position on chromosomes. Hereditas. 52(2):201–220.
- Rottenberg A, Zohary D. 1996. The wild ancestry of the cultivated artichoke. Genet Res Crop Evol. 43(1):53–58.
- Rottenberg A, Zohary D, Nevo E. 1996. Isozyme relationships between cultivated artichoke and the wild relatives. Genet Res Crop Evol. 43(1):59–62.
- Sonnante G, Carluccio AV, De Paolis A, Pignone D. 2008. Identification of artichoke SSR markers: molecular variation and patterns of diversity in genetically cohesive taxa and wild allies. Genet Res Crop Evol. 55(7):1029–1046.
- Sonnante G, Carluccio AV, Vilatersana R, Pignone D. 2007. On the origin of artichoke and cardon from the Cynara gene pool as revealed by rDNA sequence variation. Genet Res Crop Evol. 54(3):483–495.
- Sonnante G, De Paolis A, Lattanzio V, Perrino P. 2002. Genetic variation in wild and cultivated artichoke revealed by RAPD markers. Genet Resour Crop Evol. 49(3):247–252.
- Sonnante G, De Paolis A, Pignone D. 2004. Relationships among artichoke cultivars and same related wild taxa based on AFLP markers. Plant Genet Resour: Charact Util. 1(2–3):125–133.
- Sonnante G, Morgese A, Pignone D. 2012. The evolution of Cynara: diversity and domestication of artichoke and cardoon. Acta Horticult. 942:61–66.
- Sonnante G, Pignone D, Hammer K. 2007. The domestication of artichoke and cardoon: from Roman times to genomics age. Ann Bot. 100(5):1095–1100.
- Wiklund A. 1992. The genus Cynara L. (Asteraceae-Cardueae). Bot J Linn Soc. 109(1):75–123.

