?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Herein, we describe the synthesis and characterization of a mononuclear Ni complex, [Ni(mdeaH2)2](PhCO2)2 (1; mdeaH2 = N-methyldiethanolamine, PhCO2 = benzoate). The mononuclear Ni complex was characterized by FT-IR, UV-Vis, single-crystal X-ray diffraction, and its phase purity was confirmed by powder X-ray diffraction (PXRD). Magnetic data revealed antiferromagnetic interactions. Intermolecular interactions in the solid state were investigated by Hirshfeld surface analysis. Moreover, an assessment of potential inhibitory effects of this complex against rabies, rotavirus, and SARS CoV-2 was performed by a molecular docking study that suggests 1 has a strong affinity for three proteins, 6TOU, 7ZFM, and 7WC0, from the Protein Data Bank.
1. Introduction
Mononuclear transition metal complexes have attracted interest for biological applications where they may act as anticancer therapeutic agents or anti-SARS-CoV-2 agents. In addition, there is fundamental interest in the molecular magnetism of certain paramagnetic complexes [Citation1–3]. Investigations of the interactions of coordination complexes with DNA or proteins is crucial to design and discovery of effective drugs with high specificity and low general toxicity [Citation4]. DNA offers a variety of binding sites and modes for non-covalent interactions with metal complexes, such as electrostatic interactions, groove binding, and intercalation [Citation5]. Binding interactions between either DNA or Bovine serum albumin (BSA) with certain Ni(II) complexes suggest such species may be cytotoxic and bind with strong affinity [Citation6]. Molecular docking is a virtual screening technique that not only predicts the binding affinity but also the structure of the protein-target complex, which is necessary for optimization at the molecular level [Citation7]. Since the outbreak of the coronavirus pandemic (COVID-19) in 2019, the development of safe and effective anti-viral agents has become a focus of research [Citation8–10]. The emergence of new viral variants can undermine the effectiveness of current vaccines [Citation11]. Indeed, the recently identified E484K mutation in the receptor binding domain of the spike protein of a new SARS-CoV-2 strain has shown reduced vaccine efficacy [Citation12]. Along with SARS-CoV-2, rabies virus (RABV) and rotavirus are still a challenge for many countries in Asia and Africa despite significant efforts for mass vaccination in some developed countries [Citation13–15]. Our work is motivated by the need for new active molecules that may include organics, inorganic and metal complexes, to enable screening for high antiviral activities.
Complexes containing alkoxy ligands have impacted areas including magnetism, crystal engineering and catalysis due to their chelating and bridging capability, structural flexibility, forming mono- to polynuclear, molecular wheels, and high nuclearity clusters [Citation16–18]. Among them, N-alkylated diethanolamine ligands have attained attention, where their coordination behaviors can be affected by both the nature of the alkyl groups and the type of metal ions [Citation19]. N-Methyldiethanolamine (MdeaH2) is useful in synthesizing oxygen-bridged high nuclearity 3D coordination clusters and such complexes often possess interesting magnetic properties [Citation20]. Moreover, the introduction of auxiliary ligands based on carboxylic acids can help to maintain the structural topology and integrity of complexes by exhibiting unusual coordination modes. Noncovalent interactions play a crucial role in self-assembly and molecular recognition of solid-state structures. Hydrogen bonding and other dispersive interactions are the most reliable and widely used means of enforcing molecular recognition. These weak interactions between molecules play a pivotal role in biological systems and govern the physiochemical properties in the condense phase [Citation21–26].
Low molecular weight coordination complexes of late transition metals bound by MdeaH2 ligands are important because of their roles in bioinorganic chemistry and potential magnetic and catalytic applications [Citation27]. High binding affinity with DNA can be rationalized by the greater extent of H-bonding with this biomolecule. Such complexes may also display weak antiferromagnetic properties as well as antioxidant properties [Citation28].
The chemistry of Ni(II) with MdeaH2 is yet to be explored. In the present work, a Ni(II) complex was investigated using elemental analysis, FT-IR, UV–Vis, single-crystal X-ray diffraction, and magnetic studies. Weak noncovalent interactions were examined by Hirshfeld surface analysis, providing fingerprint plots (FPs), and enrichment ratios [Citation29–32]. Molecular simulations were used to assess the binding and interaction abilities of the reported compound as a starting point of the drug discovery for rabies, rotavirus, and SARS CoV-2.
2. Experimental
2.1. Materials and characterization
All starting chemicals and solvents such as N-methyldiethanolamine, NiCl2·6H2O, and sodium benzoate were commercially available and used without purification. X-ray diffraction measurements of the complex were made on a Rigaku Oxford Diffraction Super Nova E diffractometer (Rigaku Europe, Kemsing, UK) with Cu-Kα radiation from a microfocus source and corrected semi-empirically for absorption. Powder X-ray diffraction (PXRD) measurements were performed on a STOE STADI − P diffractometer. FT-IR was measured on a Bruker Alpha Platinum ATR Diamond from 4000–400 cm−1. Elemental analysis (CHN) was performed on a Perkin Elmer “Vario EL” analyzer. The optical properties were investigated on a Cary 500 scan Spectrophotometer UV-Vis-NIR from 200–800 nm at room temperature using 1 cm path length cells.
2.2. Synthesis of [Ni(mdeaH2)2](PhCO2)2 (1)
A mixture of N-methyl diethanolamine (mdeaH2) (298 mg, 2.5 mmol), NiCl2·6H2O (120 mg, 0.5 mmol), and sodium benzoate (PhCO2Na) (216 mg, 1.5 mmol) was dissolved in acetonitrile (25 mL). The solution was refluxed for 2 h followed by cooling to room temperature, filtered, and left to stand undisturbed. Light blue crystals suitable for single-crystal X-ray measurements were obtained overnight. The crystals were collected by filtration and washed with MeCN. Yield: 37% based on Ni. Elemental analyses (C, H, N) calculated (%) of compound C24H36N2NiO8: C 53.40, H 6.67, N 5.19; found: C 53.20, H 6.53, N 5.16. α: ν (cm−1): 2999(w), 2978(w), 2913(w), 2053(w), 1899(br), 1595(br), 1528(br), 1457(w), 1418(w), 1374(s), 1283(m), 1257(w), 1198(w), 1174(w), 1142(m), 1062(s), 1049(s), 1018(s), 994(s), 905(m), 882(vs), 810(s), 758(m), 714(w), 671(vs), 613(s), 554(m), 554(m), 482(m), 435(s).
2.3. X-ray Crystallography
Diffraction data for 1 was collected at 150 K on a Rigaku Oxford Diffraction Super Nova E diffractometer (Rigaku Europe, Kemsing, UK) with Cu-Kα radiation from a microfocus source. A summary of pertinent crystal data and refinement results are shown in . Using all data from the Olex 2 platform [Citation33], the structure was solved using dual-space intrinsic phasing (SHELXT) [Citation34] and refined using full-matrix least-squares refinement (SHELXL-2015) [Citation35]. Anisotropic thermal parameters were assigned to all non-H atoms, and organic H-atoms were placed in calculated positions. Crystallographic data for 2 has been deposited to the Cambridge Crystallographic Data Center, CCDC No. 2267673; copies may be obtained from www.ccdc.cam.ac.uk/conts/retrieving.html or the CCDC, 12 Union Road, Cambridge, CB2 1EZ, UK (E-mail: [email protected]).
Table 1. Crystal data, data collection, and structure refinement parameters of [Ni(mdeaH2)2](PhCO2)2.
2.4. Magnetic studies
The magnetic susceptibility measurements were obtained with the use of a Quantum Design SQUID magnetometer MPMS-XL between 1.8 and 300 K for direct current (DC) applied fields ranging from −7 to 7 T. Measurements were performed on polycrystalline samples using ∼12 mg of 1. No alternating current (AC) susceptibility peaks were found, which was checked using an oscillating AC field of 3 Oe and AC frequency of 1000 Hz. To investigate the presence of ferromagnetic impurities M versus H measurements were performed at 100 K.
2.5. Hirshfeld surface analysis
Hirshfeld surfaces analysis for 1 was carried out using CrystalExplorer 17.5 [Citation36]. A contact distance (dnorm) is based on the radii de, di, and van der Waals of the indicating atoms with color-coding by red, blue, and white on the Hirshfeld surface (defined using EquationEquation (1)(1)
(1) ), each color representing the type of contacts which depend on their distances. Red indicates the contacts with distances smaller than the total van der Waals radii (which have short contacts), white zones represent contacts with distances equal to the sum of van der Waals radii, and blue parts are free of any close contact [Citation37]. The Hirshfeld surfaces analysis of 1 was produced using CrystalExplorer 17.5 [Citation36], which can read the crystal data in CIF format.
(1)
(1)
In addition, for each point on the Hirshfeld surface, a pair distance (de, di) was expressed where de represents the distance from the point closest to the outer core and di represents the distance to the nearest inner core to the surface. The fingerprint plots (FPs) are generated by the Hirshfeld surface and the graphs of fingerprint plots are plotted by pairs (de, di) (X = di and Y = de) on the XY grid. The frequency of existence of interactions (number of points with a given pair (de, di) are represented by different colors e.g. blue, green and red showing low, medium and high frequency, respectively. Furthermore, complementary areas are observed in fingerprint plots where one molecule is a donor (de > di) and the other an acceptor (de < di) [Citation38]. Furthermore, complementary areas are observed in fingerprint plots where one molecule functions as a donor (de > di) and the other as an acceptor (de < di) [Citation38].
2.6. Molecular docking method
Docking has become an essential component in biomedical sciences as an effective strategy when the complicated 3D structure of a protein receptor is known [Citation39] and can provide a reliable model for evaluating the binding of ligands within protein targets. Protein-ligand interactions between 1 and the target proteins 6TOU, 7ZFM, and 7WC0 corresponding to the major proteases Rabies, Rotavirus, and SARS CoV-2 were determined by Autodock Vina molecular binding [Citation40]. These proteins were downloaded from a protein database (6TOU, 7ZFM, and 7WC0 PDB http://www.rcsb.org./pdb) for the binding process.
3. Results and discussion
3.1. Synthesis
To explore the coordination chemistry of 1, a self-assembly reaction at high temperature in a non-aqueous medium, between nickel chloride hexahydrate, N-methyldiethanolamine as the main ligand, and benzoic acid as a crystallization mediator, resulted in the generation of [Ni(mdeaH2)2](PhCO2)2. The phase purity of 1 was confirmed by powder X-ray diffraction (Figure S2). The product was isolated as a light blue, air-stable microcrystalline solid and was further characterized by elemental analysis, DSC-TGA, IR and UV-Vis spectroscopy, single crystal X-ray diffraction, and Hirshfeld surface analysis.
3.2. UV-Vis spectroscopy
Electronic spectral data of 1 were obtained from acetonitrile solutions recorded in the range of 200–800 nm (Figure S2). Generally, intense absorption bands are seen in the UV (λ = 220–360 nm) region and are assigned to ligand-centered π-π* transitions (which are attributed to a ligand-metal charge transfer associated with nitrogen and oxygen donors) [Citation41–45]. The absorption maxima of the bands at 226 nm (ϵ = 127,200 mol−1 L cm−1) were attributed to benzoate-based π-π* transitions and weaker bands at 272 nm (ϵ = 29,000 mol−1 L cm−1) were attributed to an n-π* the transition of the carboxylate group.
3.3. Thermal stability
Thermal gravimetric analysis of 1 was carried out to confirm composition and to understand the thermal stability (Figure S4). The Ni(II) complex was heated from 30–1000 °C under nitrogen. The TGA curve displayed two steps of weight loss. The first weight loss was 40% from 220 to 311 °C, which corresponds to removal of the two benzoate molecules present in the crystal structure (theoretical value is 44.92%). The two benzoate molecules within the crystal structure is depicted in . Smooth TGA curves showed that the two benzoate molecules have similar characteristics. The second thermal degradation reaction occurred in a single step between 311–386 °C with a weight loss of 44.00%, in agreement with loss of two organic ligands (m-deaH2) (calculated value is 44. 18%). The injection of O2 at 390 °C caused the removal of the organic part and the formation of nickel oxide (NiO). The residual NiO amounts match their theoretical values [Citation43, Citation46].
3.4. X-ray crystallography
Single crystal X-ray diffraction studies reveal that 1 is a mononuclear entity that crystallizes in the monoclinic space group P21/c with Z = 2 as shown in . The crystallographic data and structure refinement parameters for 1 are given in , and selected bond distances and angles are in . 1 is a centrosymmetric complex with an inversion center. The primary ligand, Me-deaH2, is coordinated to nickel ion through its two nitrogen and four oxygen atoms in a neutral form with (η1: η1: η1: μ1) coordination. The bond length of metal to nitrogen of the ligand is Ni-N1 and Ni-N1′ = 2.098(1) Å and bond distances of nickel to oxygen of the ligand Ni-O(1) and Ni-O(1)′ = 2.075(1) and Ni-O(2) and Ni-O(2)′ = 2.061(1) Å. The structure suggests that nickel is in a + 2 oxidation state which is stabilized by two counter-anions of benzoate. The bond angles N1–Ni1–N1, O4–Ni1–O4, and O5–Ni1–O5 are aligned at 180° as shown in . The atoms of the molecule adjacent to each other interact through non-covalent interactions with several short contacts observed. This type of interaction is seen as O–H···C = 2.667 Å and hydrogen bonding is seen between the hydrogen of the main ligand and the oxygen of the benzoate anion as O–H···O contacts with bond distances 1.62 Å and 1.64 Å and C-H···π interactions (3.846 Å) resulting in a 1D chain in Figure S1. Two benzoate anions surround the metal and ligand to form [Ni(mdeaH2)2](PhCO2)2. The distorted octahedral geometry of NiN2O4 was confirmed by SHAPE analysis [Citation47–50] with a deviation of 0.59 (Table S1).
Table 2. Selected bond lengths and angles of 1.
3.5. Magnetic measurements
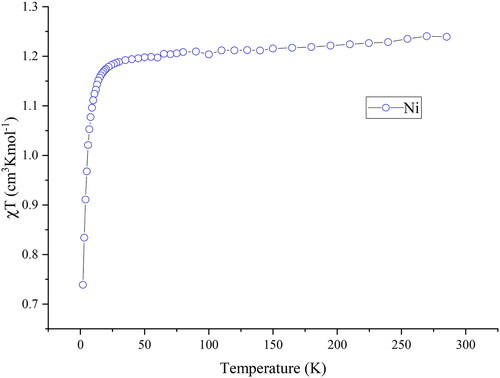
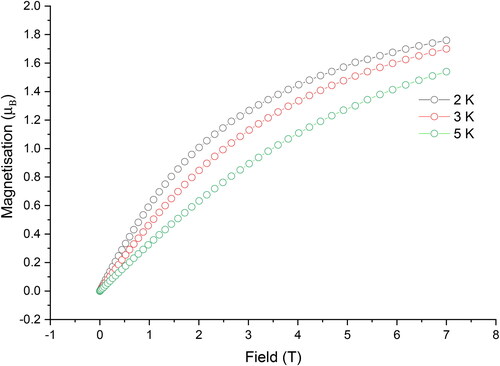
Magnetic measurements on 1 were conducted using a Quantum Design MPMS-XL SQUID magnetometer with magnets capable of applying DC fields up to 7 T. The DC measurement of 1 was carried out on a fresh microcrystal sample from 2–300 K under an applied DC magnetic field of 1000 Oe (0.1 T); the plot of χT versus T for 1 is shown in . The χMT value is 1.24 cm3 K mol−1 at room temperature, which is close to expected for one isolated NiII ion (1.28 cm3 mol−1 K for g = 2.0 and S = 1) for a mononuclear nickel(II) complex [Citation51,Citation52]. The value of χMT slightly decreases upon cooling between 300 to 50 K followed by a rapid decrease from 50 to 2 K, reaching a value of 0.74 cm3 mol−1 K at 2 K, further confirming the strong antiferromagnetic interaction. The magnetization of 1 was measured at fields ranging from 0 to 70000 Oe (0–7 T) at temperatures of 2 K, 3 K, and 5 K. shows the magnetization values of 1 have a relatively rapid increase below 1 T followed by increase linearly up to 7 T, reaching a value of 1.76 µB at 2 K and 7 T without saturation, indicating the presence of magnetic anisotropy or/and the population of low-lying excited states [Citation53].
3.6. Hirshfeld surface analysis
The analysis of Hirshfeld surfaces provides additional insight into weak intermolecular interactions which are influential in the packing of molecules in crystals. A Hirshfeld surface is defined as the density weight function of the specific molecule (i.e. the pro-molecule) over the same sum of density of its nearest neighbor (i.e. the pro-crystal), thereby resulting in a 0.5 arbitrary units iso surface, which is similar to that of a van der Waals surface but, unlike the latter, takes into consideration neighboring molecules and hence provides information about intermolecular interactions [Citation54]. The Hirshfeld surfaces can be mapped with diverse properties, dnorm, electrostatic potential, shape-index and curvedness. These are useful to accumulate additional information on weak intermolecular interactions.
The Hirshfeld surfaces mapped over dnorm employ the function of normalized distances de and di, where de and di are the distances from a given point on the surface to the nearest atom outside and inside, respectively. The blue, white and red colors used for the dnorm-mapped Hirshfeld surfaces recognize the interatomic contacts as longer, at van der Waals separations and short interatomic contacts, respectively. The useful measures of curvature, namely curvedness and shape-index, introduced by Koendrink provide further chemical insight into molecular packing [Citation55]. A surface with low curvedness may indicate π–π stacking in the crystal. On the other hand, a Hirshfeld surface with high curvedness is designated as dark-blue edges, indicative of absence of π–π stacking. The shape-index is a qualitative measure of shape and is sensitive to subtle changes in surface shape, predominantly in a flat region. Two shape indices differing by sign represent complementary “bumps and hollows.” The blue bump-shape and shape-index > 1 belongs to the donor and that representing a red hollow with index < 1 corresponds to the acceptor of an intermolecular interaction.
The two-dimensional fingerprint plots derived from a Hirshfeld surface offer a visual summary of the frequency of each combination of de and di across the surface of a molecule. The color of each point corresponding to the relative area of a (de, di) pair is recognized as the contribution from different interatomic contacts: blue, green and red correspond to small, moderate and greatest contributions whereas an uncolored region indicates no contribution to the Hirshfeld surface. A fingerprint plot outlined into specific interatomic contacts contains information related to specific intermolecular interactions.
The Hirshfeld surface analysis for 1 revealed surfaces mapped over the dnorm range of 0.5–1.5 Å for investigation of further supramolecular non-covalent interactions. The white spots indicate contacts with distances equal to the sum of van der Waals radii and the red color depressions indicate non-covalent contacts visible on the transparent dnorm surfaces. The molecular Hirshfeld surface analyses for 1 are mapped over the dnorm range −0.807 to 1.323 Å, shape index range −0.998 to 0.998 Å, curvedness range −3.341 to 0.294 Å, di range 0.603–2.583 Å and de range 0.970–2.558 Å (). Strong O···H intermolecular interactions indicated by dnorm mapping exist between nitrogen and hydrogen of the coordinating ligand (O···H = 2.79 Å).
Figure 4. Molecular Hirshfeld surfaces (a) dnorm, (b) shape index, (c) curvedness, (d) di and (e) de of 1.
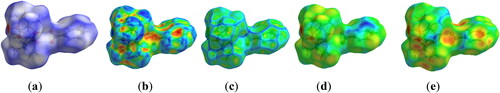
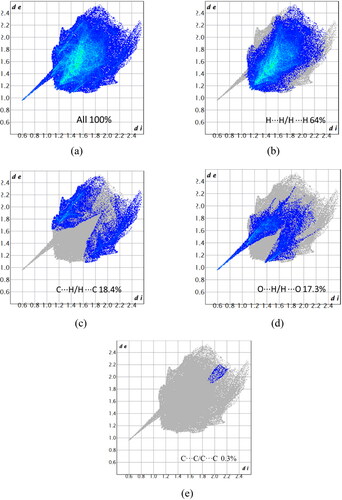
The intermolecular H···H, O···H, and C···H interactions complement the Hirshfeld surfaces generated by the 2D fingerprint plots. The fingerprint plots show particular atom-pair close contacts and enables the separation of contributions from different types of interactions that overlap in the full fingerprint [Citation56]. Furthermore, it also reveals that one molecule acts as a donor (de > di) and the other as an acceptor (de < di) visible in a complementary region. Many short contacts are revealed in the fingerprint plot (). The broad region bearing short and narrow spikes in the middle of the plot is reflected as H–H/H–H interaction comprising 64% interaction of the iron complex, as depicted in . Two sharp spikes pointing toward the lower left of the plot is typical for C–H/H–C comprising 18.4% interactions (). This portion corresponds to O–H/H–O interactions comprising 17.3% () and 0.3% for C-C interactions of the given complex ().
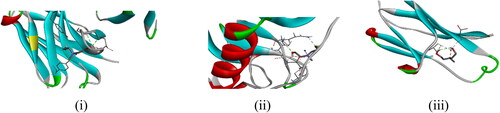
3.7. Molecular docking
The theoretical protein binding modes of 1 were investigated by molecular docking (). The proteins 6TOU, 7ZFM, and 7WC0 are linked with viral diseases like rabies, rotavirus, and SARS CoV-2. The binding energies, inhibition constants, and amino acid residues that link with 1 and their bond distances are shown in . Specific protein sites were identified for each biomolecule. For instance, amino acid residues for the protein 6TOU are TYR52, SER98, GLY100, TYR1094, and TRY52, for the protein 7ZFWM the amino acid residues are GLN102, THR100 and SER98 and for the protein 7WC0 the amino acid residues are SER174 and THR196.
Table 3. The binding energies, inhibition constant, amino acid residues, and bond distances linked with 1.
4. Conclusion
A new mononuclear complex, [Ni(mdeaH2)2](PhCO2)2, was synthesized from N-methyldiethanolamine with benzoic acid as a co-ligand. 1 crystallized in a monoclinic space group and exhibited a distorted octahedral geometry which was confirmed by DC magnetism data where a χMT value of 1.24 cm3 K mol−1 was found at room temperature, close to what is expected for an isolated NiII ion (1.28 cm3 mol−1 K for g = 2.0 and S = 1). The absorbance spectra of 1 provided further information about electronic structure. The absorption maxima of the bands found at 226 nm (ϵ = 127,200 mol−1 L cm−1) were attributed to π-π* transitions in benzoate and the weaker band at 272 nm (ϵ = 29,000 mol−1 L cm−1) was attributed to an n-π* transition of the carboxylate. The strong O···H bond intermolecular interactions indicated by dnorm mapping exist between nitrogen and hydrogen of the coordinating ligand (O···H = 2.79 Å). It further revealed that 1 has been stabilized mainly by H···H, C···H/H···C, O···H/H···O contacts. Molecular docking was carried out to investigate the interactions of 1 with proteins of SARS-CoV-2, rabies, and rotavirus, indicating strong binding affinities.
Supplemental Material
Download MS Word (731.3 KB)Acknowledgements
The author G. Abbas acknowledges Prof. Annie K. Powell for providing the lab facilities at KIT, Karlsruhe, Germany, and further extension of studies funded by George Forster Fellowship by Alexander von Humboldt Foundation Germany.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Q. Umar, Y. Huang, A. Nazeer, H. Yin, J. Zhang, M. Luo, X. Meng. RSC Adv., 12, 32119 (2022).
- K. Ioannou, M.C. Vlasiou. Biometals, 35, 639(2022).
- J. Miklovič, D. Valigura, R. Boča, J. Titiš. Dalton Trans., 44, 12484 (2015).
- J. Haribabu, K. Jeyalakshmi, Y. Arun, N.S. Bhuvanesh, P.T. Perumal, R. Karvembu. J. Biol. Inorg. Chem., 22, 461 (2017).
- A.M. Abu-Dief, R.M. El-Khatib, F.S. Aljohani, S.O. Alzahrani, A. Mahran, M.E. Khalifa, N.M. El-Metwaly. J. Mol. Struct., 1242, 130693 (2021).
- M. Niu, Z. Li, H. Li, X. Li, J. Dou, S. Wang. RSC Adv., 5, 37085 (2015).
- X.-Y. Meng, H.-X. Zhang, M. Mezei, M. Cui. Curr. Comput. Aided Drug Des., 7, 146 (2011).
- G. Abbas, A. Irfan, I. Ahmed, F.K. Al-Zeidaneen, S. Muthu, O. Fuhr, R. Thomas. J. Mol. Struct., 1253, 132242 (2022).
- J. Karges, S.M. Cohen. ChemBioChem., 22, 2600 (2021).
- Niaz, Muhammad, Ishaq N, Khan, Zafar, Ali, Mohammad, Ibrahim, Shaukat, Shujah, Saqib, Ali, Muhammad, Ikram, Sadia, Rehman, Gul Shahzada, Khan, Abdul, Wadood, Awal, Noor, Carola, Schulzke, Viola. J. Mol. Struct., 1253, 132308 (2022).
- Y. Weisblum, F. Schmidt, F. Zhang, J. DaSilva, D. Poston, J.C. Lorenzi, F. Muecksch, M. Rutkowska, H.-H. Hoffmann, E. Michailidis, C. Gaebler, M. Agudelo, A. Cho, Z. Wang, A. Gazumyan, M. Cipolla, L. Luchsinger, C.D. Hillyer, M. Caskey, D.F. Robbiani, C.M. Rice, M.C. Nussenzweig, T. Hatziioannou, P.D. Bieniasz. Elife, 9, e61312 (2020).
- N.A. Muena, T. García-Salum, C. Pardo-Roa, E.F. Serrano, J. Levican, M.J. Avendaño, L.I. Almonacid, G. Valenzuela, E. Poblete, S. Strohmeier, E. Salinas, D. Haslwanter, M.E. Dieterle, R.K. Jangra, K. Chandran, C. González, A. Riquelme, F. Krammer, N.D. Tischler, R.A. Medina. medRxiv, (2021).
- H. Bourhy, A. Dautry-Varsat, P.J. Hotez, J. Salomon. Rabies, still neglected after 125 years of vaccination, Public Library of Science San Francisco, USA (2010).
- J. Hellert, J. Buchrieser, F. Larrous, A. Minola, G.D. de Melo, L. Soriaga, P. England, A. Haouz, A. Telenti, O. Schwartz, D. Corti, H. Bourhy, F.A. Rey. Nat. Commun., 11, 596 (2020).
- S.E. Crawford, S. Ramani, J.E. Tate, U.D. Parashar, L. Svensson, M. Hagbom, M.A. Franco, H.B. Greenberg, M. O’Ryan, G. Kang, U. Desselberger, M.K. Estes. Nat. Rev. Di.s Primers, 3, 17083 (2017).
- E.M. Rumberger, S.J. Shah, C.C. Beedle, L.N. Zakharov, A.L. Rheingold, D.N. Hendrickson. Inorg. Chem., 44, 2742 (2005).
- K.R. Gruenwald, A.M. Kirillov, M. Haukka, J. Sanchiz, A.J. Pombeiro. Dalton Trans., 12, 2109 (2009).
- D. Schray, G. Abbas, Y. Lan, V. Mereacre, A. Sundt, J. Dreiser, O. Waldmann, G.E. Kostakis, C.E. Anson, A.K. Powell. Angew. Chem. Int. Ed. Engl., 49, 5185 (2010).
- G. Abbas, Y. Lan, V. Mereacre, G. Buth, M.T. Sougrati, F. Grandjean, G.J. Long, C.E. Anson, A.K. Powell. Inorg. Chem., 52, 11767 (2013).
- T. Nakajima, K. Seto, F. Horikawa, I. Shimizu, A. Scheurer, B. Kure, T. Kajiwara, T. Tanase, M. Mikuriya. Inorg. Chem., 51, 12503 (2012).
- S.K. Seth, I. Saha, C. Estarellas, A. Frontera, T. Kar, S. Mukhopadhyay. Cryst. Growth Des., 11, 3250 (2011).
- S.K. Seth, P. Manna, N.J. Singh, M. Mitra, A.D. Jana, A. Das, S.R. Choudhury, T. Kar, S. Mukhopadhyay, K.S. Kim. CrystEngComm, 15, 1285 (2013).
- S. Tripathi, S. Islam, S.K. Seth, A. Bauzá, A. Frontera, S. Mukhopadhyay. CrystEngComm, 22, 8171 (2020).
- S.K. Seth, A. Bauzá, A. Frontera. New J. Chem., 42, 12134 (2018).
- S.K. Seth, A. Bauzá, G. Mahmoudi, V. Stilinović, E. López-Torres, G. Zaragoza, A.D. Keramidas, A. Frontera. CrystEngComm, 20, 5033 (2018).
- G. Abbas, F.C. Franco, S. Janani, J.C. Mishma, A. Manikandan, S. Muthu, C.E. Anson. J. Mol. Struct., 1297, 136933 (2023).
- F. Sama, A.K. Dhara, M.N. Akhtar, Y.-C. Chen, M.-L. Tong, I.A. Ansari, M. Raizada, M. Ahmad, M. Shahid, Z.A. Siddiqi. Dalton Trans., 46, 9801 (2017).
- M. I, M. Shahid, M. Ahmad, R. Rahisuddin, R. Arif, S. Tasneem, F. Sama, I.A. Ansari. New J. Chem., 43, 622 (2019).
- S.K. Seth, A. Bauzá, A. Frontera. CrystEngComm, 20, 746 (2018).
- S.K. Seth. Acta Crystallogr E Crystallogr Commun, 74, 600 (2018).
- S.K. Seth. Crystals, 8, 455 (2018).
- S. Tripathi, A. Hossain, S.K. Seth, S. Mukhopadhyay. J. Mol. Struct., 1226, 129254 (2021).
- O.V. Dolomanov, L.J. Bourhis, R.J. Gildea, J.A. Howard, H. Puschmann. J. Appl. Crystallogr., 42, 339 (2009).
- G.M. Sheldrick. Acta Crystallogr. A Found. Adv., 71, 3 (2015).
- G.M. Sheldrick. Acta Crystallogr. C Struct. Chem., 71, 3 (2015).
- M. Turner, J. McKinnon, S. Wolff, D. Grimwood, P. Spackman, D. Jayatilaka, M. Spackman. CrystalExplorer (Version 17.5), University of Western Australia, Crawley, Australia (2017).
- S.K. Seth, N.C. Saha, S. Ghosh, T. Kar. Chem. Phys. Lett, 506, 309 (2011).
- M.A. Spackman, J.J. McKinnon. CrystEngComm, 4, 378 (2002).
- I.D. Kuntz, J.M. Blaney, S.J. Oatley, R. Langridge, T.E. Ferrin. J. Mol. Biol., 161, 269 (1982).
- J. Eberhardt, D. Santos-Martins, A.F. Tillack, S. Forli. J. Chem. Inf. Model, 61, 3891 (2021).
- T.M. Donlevy, T.W. Hambley, G.R. Hanson, K.L. McMahon, R. Stranger, L.R. Gahan. Inorg. Chem., 33, 5131 (1994).
- R. Cao, Q. Shi, D. Sun, M. Hong, W. Bi, Y. Zhao. Inorg. Chem., 41, 6161 (2002).
- I.-T. Lim, C.-H. Kim, K.-Y. Choi. J. Korean Chem. Soc., 62, 427 (2018).
- A. Saeed, A.U. Rahman, N.U. Islam, M. Saeed, S.A. Halimi. Malay. J. Pharm. Sci., 4, 63 (2006).
- C. Lund Myhre, C.J. Nielsen. Atmos. Chem. Phys., 4, 1759 (2004).
- M. Taştekin, S. Durmuş, E. Şahin, C. Arici, K.C. Emregül, O. Atakol. Z. Kristallogr. Cryst. Mater, 223, 424 (2008).
- M. Jones. Design and Synthesis of Novel Lanthanide Complexes for Luminescent Devices, Cardiff University, United Kingdom (2006).
- C. Polyzou. … Σχολή Θετικών Επιστημών. Τμήμα Χημείας. Εργαστήριο, (2016).
- A. Abragam, B. Bleaney. Electron Paramagnetic Resonance of Transition Ions, OUP Oxford, United Kingdom (2012).
- L. Sorace, C. Benelli, D. Gatteschi. Chem. Soc. Rev., 40, 3092 (2011).
- B. Zhai, Z.Y. Li, F.Q. Zhang, S.Z. Li, F.L. Zhang, C. Zhang, X.F. Zhang, G.X. Cao, Y. Ma. Zeitschrift Anorg Allge Chemie, 642, 527 (2016).
- N. Zhang, Y.-J. Sun, H. Yang, J.-Y. Zhang, E.-Q. Gao. Inorg. Chim. Acta, 428, 37 (2015).
- D.I. Alexandropoulos, L. Cunha-Silva, L. Pham, V. Bekiari, G. Christou, T.C. Stamatatos. Inorg. Chem., 53, 3220 (2014).
- J.J. McKinnon, D. Jayatilaka, M.A. Spackman. Chem. Commun. (Camb), 37, 3814 (2007).
- J.J. Koenderink. Solid Shape, MIT Press, USA (1990).
- I.A. Ansari, F. Sama, M. Raizada, M. Shahid, M. Ahmad, Z.A. Siddiqi. New J. Chem., 40, 9840 (2016).

2. Color code: black, red, blue, turquoise, and white spheres represent C, O, N, Ni, and H, respectively.](/cms/asset/706be66f-641b-4bd4-9323-8b9b2095406d/gcoo_a_2318779_f0001_c.jpg)
2 (1).](/cms/asset/3bd4a58d-9708-487a-aa63-f81e4bf5e4ab/gcoo_a_2318779_sch0001_c.jpg)