Abstract
Parental experience can modulate the behavior of their progeny. While the molecular mechanisms underlying parental effects or inheritance of behavioral traits have been studied under several environmental conditions, it remains largely unexplored how the nature of parental experience affects the information transferred to the next generation. To address this question, we used C. elegans, a nematode that feeds on bacteria in its habitat. Some of these bacteria are pathogenic and the worm learns to avoid them after a brief exposure. We found, unexpectedly, that a short parental experience increased the preference for the pathogen in the progeny. Furthermore, increasing the duration of parental exposure switched the response of the progeny from attraction to avoidance. To characterize the underlying molecular mechanisms, we found that the RNA-dependent RNA Polymerase (RdRP) RRF-3, required for the biogenesis of 26 G endo-siRNAs, regulated both types of intergenerational effects. Together, we show that different parental experiences with the same environmental stimulus generate different effects on the behavior of the progeny through small RNA-mediated regulation of gene expression.
Introduction
In many organisms, the behaviour and physiology of progeny are modulated by parental experience, ultimately generating traits that often resemble the acquired phenotype of the parents (Bohacek & Mansuy, Citation2015; Dias, Maddox, Klengel, & Ressler, Citation2015; Horsthemke, Citation2018; Miska & Ferguson-Smith, Citation2016; Weigel & Colot, Citation2012). A growing body of evidence documents the intergenerational transfer of acquired traits, such as stress response or olfactory sensitivity in rodents (Dias & Ressler, Citation2014; Gapp et al., Citation2014). This form of plasticity “anticipates” that the next generation will experience a similar environment to their parents, in which case it may prove to be adaptive. However, the same parental experience can affect progeny differentially depending on their sex, genotype, further ancestral history and life experience (Bohacek & Mansuy, Citation2015; Deas, Blondel, & Extavour, Citation2019; Kundakovic et al., Citation2013; Palominos et al., Citation2017). These observations suggest that inter- and transgenerational regulations are influenced by many different factors and do not always copy the parentally acquired traits.
Despite our increased knowledge on transgenerational effects, the impact of duration, intensity or frequency of parental experience on modulating certain traits in their progeny remains largely unexplored. Here, we examine this question using C. elegans, a nematode that feeds on bacteria of different genuses. The chemotactic behaviour of C. elegans towards this food source is highly dependent on the olfactory system, which is critical for detecting food and avoiding dangers. Bacteria from the Pseudomonas genus are abundant in the natural habit of the nematode (Félix & Duveau, Citation2012; Samuel, Rowedder, Braendle, Félix, & Ruvkun, Citation2016), but certain Pseudomonas strains, such as Pseudomonas aeruginosa PA14, infect the worm after being ingested, which results in a slow death over several days (Tan, Mahajan-Miklos, & Ausubel, Citation1999). Therefore, Pseudomonas aeruginosa PA14 is both food and pathogen to C. elegans and plays a critical role in the survival of the worm.
We have previously shown that adult C. elegans robustly learns to reduce their preference for the odorants of PA14 after transiently feeding on the pathogen for 4 h (Ha et al., Citation2010; Zhang, Lu, & Bargmann, Citation2005). This form of learning is contingent on the pathogenesis of the bacteria and resembles the Garcia effect, a form of conditioned aversion that allows animals to learn to avoid the smell or taste of a food that makes them ill (Garcia, Hankins, & Rusiniak, Citation1974; Ha et al., Citation2010; Zhang et al., Citation2005). Because continuously ingesting PA14 can be lethal to the worm, intergenerational transfer of information regarding the valence of the cues that signal this food source could be adaptive for the next generation. Therefore, we asked whether learning to avoid PA14 in C. elegans mothers regulated the olfactory response to PA14 in progeny. Furthermore, we asked whether parental learning experience of different durations differentially affected the behavioral response of the progeny.
To elucidate the molecular pathways underlying parental experience-induced behavioral changes in progeny, we characterized the small non-coding RNA pathways previously implicated in epigenetic inheritance in different species (Bohacek & Mansuy, Citation2015; Gapp et al., Citation2014; Miska & Ferguson-Smith, Citation2016; Moore et al., Citation2019; Okamura & Lai, Citation2008; Palominos et al., Citation2017; Posner et al., Citation2019; Rechavi et al., Citation2014). Three major classes of small RNAs have been described in C. elegans: small interfering RNAs (endo-siRNAs and exo-siRNAs for endogenously and exogenously produced small interfering RNAs, respectively), microRNAs and PIWI-interacting RNAs (piRNAs). Endo-siRNAs are encoded in the genome and target worm transcripts; the RNA-dependent RNA Polymerase (RdRP) RRF-3 uses endogenous mRNAs as templates to synthesize the complementary strand. The resulting double-stranded RNAs (dsRNAs) are cleaved into 26 G endo-siRNAs that target genes expressed in the soma and germline. Mutating RRF-3 downregulates 26 G endo-siRNAs in the sperm, oocytes and embryos and upregulates the expression of many genes (Gent et al., Citation2010; Han et al., Citation2009; Lee, Hammell, & Ambros, Citation2006; Simmer et al., Citation2002). The piRNAs are 21 to 22-nucleotide RNAs enriched in the germline where they protect the integrity of the genome and regulate germ cell development and fertility. The piRNAs form complexes with the PIWI-clade Argonautes, such as PRG-1, to target both endogenous and exogenous sequences (Ashe et al., Citation2012; Batista et al., Citation2008; Das et al., Citation2008; Lee et al., Citation2012; Wang & Reinke, Citation2008)) . Given the documented role of these two pathways in epigenetic inheritance, we characterized the function of RRF-3 and PRG-2 in aversive learning of PA14 in C. elegans adults and the effects of the parental learning experience on the olfactory response of the progeny. Our results show that parental learning experience of different durations differentially modulate the behavior of the progeny and that the endo-siRNA pathways regulate these intergenerational effects.
Materials and methods
Experimental models and subject details
C. elegans hermaphrodites were used in this study and cultivated at 20 °C under standard conditions (Brenner, Citation1974). The strains used in this study include: N2 (Bristol), ZC2834 rrf-3(pk1426) II, YY13 rrf-3(mg373) II, ZC2987 prg-2(n4358) IV, WM162 prg-2(tm1094) IV, CX4998 kyIs140 I; nsy-1(ky397) II.
Bacterial strains
For this study we used the non-pathogenic bacteria Escherichia coli OP50 and the pathogenic bacteria Pseudomonas aeruginosa PA14.
Method details
Aversive olfactory training of P0s with PA14 and cultivation of F1s
Training of P0s (mothers) with PA14 was performed as previously described (Ha et al., Citation2010; Zhang et al., Citation2005) with minor modifications. Individual OP50 or PA14 colonies were inoculated in 50 mL nematode growth medium (NGM) and cultivated with shaking at 27 °C overnight. 800 µL (for 4-h training) or 400 µL (for 8-h training) of OP50 or PA14 culture was spread onto 10 cm NGM plate and incubated at 27 °C for 2 days to prepare the naive control plates and the training plates, respectively. Tests performed prior to the beginning of the experiments showed that 800 µL-seeded plates generated a very stable learning after 4-h exposure. However, the same amount of bacteria seeded in training plates used for 8-h training caused severe locomotor impairment in P0s that could not be tested in the droplet assay. Therefore, we used 400 µL PA14 culture to seed the training plates for 8-h training.
P0 eggs were first extracted from 1-day adult hermaphrodites cultivated under standard conditions (Brenner, Citation1974) and grown until they reached adulthood. Synchronized young adult P0s were then transferred to the training (PA14 seeded) or the control (OP50 seeded) plate and kept at 20 °C for 4h or 8 h depending on the experimental procedure. By the end of the training, some naive control and trained P0 worms were randomly picked from the plates and tested in parallel for their olfactory choice and learning in the droplet assay (see next section for details). The rest of the P0 worms were collected with S-basal medium and passed through a cell strainer 40-micrometer nylon filter (Falcon) to remove laid eggs. The P0 worms were then treated with a bleach solution to isolate F1 embryos, which were hatched and cultivated on plates seeded with an OP50 culture grown overnight in LB medium and kept at 20 °C until the adult stage.
Olfactory preference assay using the automated olfactory assay
The automated assay (Droplet assay) that quantifies olfactory preference in individual animals was performed as described [(Ha et al., Citation2010) and ] with some modifications. Briefly, after training, 6 naive and 6 trained P0s per assay were randomly picked, washed, and individually placed in the droplets of 2 µL NGM buffer in an enclosed chamber. Worms were exposed to alternating airstreams odorized with OP50 or PA14 by blowing clean air through the supernatant of freshly generated bacterial cultures. Each olfactory stimulation lasted for 30 s and each assay contained 12 cycles of stimulation. Naive and trained worms were tested in parallel. The locomotion of the worms was recorded and the large body bends (Ω bends, i.e. body bends in the shape of Ω) of the tested worms were quantified with computational analysis performed in Matlab. Because Ω bends are followed by reorientation, a higher rate of Ω bends indicates a lower preference for the tested airstream (Ha et al., Citation2010). The P0 Choice Index (CI) of each worm was defined as the turning rate evoked by OP50 smell minus the turning rate evoked by PA14 smell and normalized by the sum of the rates (). The CI of each assay was quantified as the median value of the CIs of the individual P0 worms in that assay. Each test session consisted of two assays (with a total of 12 naive and 12 trained worms being tested). Individual data points in the graphs represent the average CI of the two assays of the test session. If technical problems occurred during testing, the affected assay was excluded from analysis. Any worm that was injured during the process of transferring to the testing chamber was also excluded. The P0 Learning Index (LI) of each session was defined as the CI of the naive worms (P0 OP50) minus the CI of the trained worms (P0 PA14) (). Individual data points in the graphs represent the average LIs of the two assays of individual test sessions. A positive LI indicates learned avoidance of PA14. The total number of worms per experiment is specified in Supplemental Table 1.
Figure 1. Experiment design to examine effect of parental experience with PA14 on progeny. Schematics for the training procedure and the learning assays.
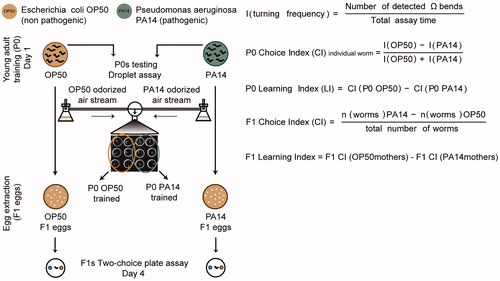
Importantly, the droplet assay allowed us to test several P0s simultaneously, with minimal disturbance of the other P0s left on the plate, whose embryos (F1s) were collected for testing.
Olfactory preference assay using the two-choice assay
The two-choice plate assay is similar to the one previously described (Zhang et al., Citation2005), except for several modifications. To measure the olfactory preference for bacteria odorants, a drop of 5 µL supernatant of OP50 and a drop of 5 µL supernatant of PA14 freshly generated bacterial cultures were put 2 cm apart on a 6 cm NGM plate. In each assay, one worm was placed on the plate equidistant to the two drops of the culture supernatant right after the drops were put on the plate and allowed to crawl to the preferred stimulus.
For each assay 8 worms were tested per condition. The worms that did not make a choice after 10 min were counted in the total number. The worms that disappeared during the assay by moving outside of the plate were not included. The Choice Index in the two-choice assay was defined as the number of worms that chose PA14 minus the number of worms that chose OP50 normalized by the total number of worms tested (). When testing the progeny, the F1 Learning Index for the two-choice assay was the Choice Index of F1 progeny from naive P0s (OP50mothers) minus the Choice Index of F1 progeny from trained P0s (PA14mothers) (). A positive F1 Learning Index indicates an increased avoidance of PA14 in F1s induced by the parental experience with PA14. Although we name this variable Learning Index, no F1 worm was exposed to PA14 training plates at any time. The total number of worms per experiment is shown in Supplemental Table 1.
The behavior of the F1 worms was recorded during the two-choice assay and specific behavioral features were quantified (Liu et al., Citation2018), which allowed us to examine the odorant-guided movements of the progeny in detail. The analysis was performed with the Wormlab tracker (https://www.mbfbioscience.com/wormlab) and further analysed in Matlab. While the two-choice plate assay allowed us to analyse odorant-guided chemotactic movements, it tested one worm each time. In comparison, the droplet assay tested several worms simultaneously within a few minutes, allowing us to quickly sample the learning of the hermaphrodite mothers before isolating F1 embryos.
Slow killing assay
The plates for the slow killing assay were prepared as previously described (Tan et al., Citation1999). Twenty young adult hermaphrodites were transferred onto each slow killing plate and kept at 25 °C. The dead and the live worms were counted at the specific time points as shown in the figure.
The analysis of parental effects induced by 8-h training
To analyse the parental effects induced by 8-h training, we plotted the values of F1 Learning Index as a function of the values of the Learning Index of their respective P0s and tested for a significant linear correlation between the variables using a Permutation test where the P0 and F1 learning indexes were randomly paired for 5000 times. We then calculated the average P0 Learning Index and separated the experiments into two groups—the experiments in which the value of the P0 Learning Index was higher than the average Learning Index, and the experiments in which the P0 Learning Indexes were lower than the average Learning Index ().
Quantification and statistical analysis
All statistical data analyses were conducted using Matlab_R2015b. In graphical representations each data point represents a biological replicate (n = number of biological replicates) unless stated otherwise. Data from different groups presented in the same graph (such as trained and control groups, WT and mutants) were collected in parallel. When data were normally distributed, we used parametric tests for comparison between groups and data were represented as average ± sem. When data were not normally distributed, non-parametric tests were used and data were graphically represented as median with first and third quartiles. The statistical methods, sample size and number of the replicates are shown in Supplemental Table 1.
Results
Parental exposure to PA14 for 4 h increases preference for pathogen in progeny
To examine how parental experience modulates offspring behavior, we exposed C. elegans to the pathogenic bacteria Pseudomonas aeruginosa PA14 and asked whether maternal aversive learning experience altered the olfactory response to PA14 in progeny.
We trained adult hermaphrodite mothers (P0s) on PA14 for 4 h, while in parallel the naive controls were fed with OP50 (, Materials and methods). Naive C. elegans worms prefer the odorants of PA14 in comparison with OP50, and previous results in our lab have shown that 4-h exposure to PA14 reduces this olfactory preference (Ha et al., Citation2010). Therefore, we employed this experimental model to study how an experience that changed the behavior in mothers affected the behavior of the next generation.
First, we quantified the preference between the smell of OP50 and the smell of PA14 in naive and trained mothers using the droplet assay, and consistent with our previous findings (Ha et al., Citation2010; Zhang et al., Citation2005), mothers trained with PA14 (P0 WT PA14) showed a reduced preference for the odorants of the pathogen in comparison with naive mothers (P0 WT OP50). This difference is reflected in a positive P0 Learning index, indicating that training with PA14 in P0s reduces the preference for PA14 in comparison with naive controls (, data on the left side of the graphs depicted by empty circles). We also analyzed the preference of P0s using the two-choice assay on plate (see Material and methods) and found similar training effects (Supplemental Figure 1(A)), indicating that both the droplet assay and the two-choice assay can be used to measure olfactory preference between two bacterial food sources.
Figure 2. Training with pathogenic PA14 for 4 h increases offspring olfactory preference for PA14, in a small RNA pathway dependent manner. (A,B) Naive wild-type (WT) animals fed on E. coli OP50 (P0 WT OP50) prefer the odorants of PA14 (A), and training with PA14 (P0 WT PA14) decreases the preference (A,B). The deletion mutation rrf-3(pk1426) does not alter PA14 aversive learning in P0 mothers (A,B). n = 9 biological replicates each, droplet assay. (C,D) Progeny of WT trained mothers (F1 WT PA14mothers) show an increased preference for PA14 compared with progeny of WT naive mothers (F1 WT OP50mothers). The rrf-3(pk1426) deletion abolishes this increase (C), which produces a F1 Learning Index different from WT (D). n = 9 biological replicates each, two-choice assay. (E,F) The prg-2(n4358) mothers (P0s) learn to avoid PA14 similarly to WT. Further comparisons also show that CI of P0 WT OP50 is different from CI of P0 prg-2(n4358) OP50 (p < 0.01) and CI of P0 WT PA14 is different from CI of P0 prg-2(n4358) PA14 (p < 0.01). n = 8 biological replicates each, droplet assay. (G,H) The learning experience of P0s does not alter the olfactory preference in prg-2(n4358) F1s (G), which display defective F1 Learning Index in comparison with WT (H), n = 8 biological replicates each, two-choice assay. (I,J) The rrf-3(mg373) and prg-2(tm1094) mutant mothers learn similarly to WT (I, droplet assay) but show a defective F1 Learning Index (J, two-choice assay). n = 8, 6 and 7 biological replicates for WT, rrf-3(mg373) and prg-2(tm1094), respectively. For all, ***p < 0.001, **p < 0.01, *p < 0.05. Data are normally distributed in A, B, D–F, H–J and presented with Mean ± SEM; data are not normally distributed in C, G and presented with box plots (showing median, first and third quartile, whiskers extending to values within 2.7 standard deviations). More details in statistical analyses are in Supplemental Table 1.
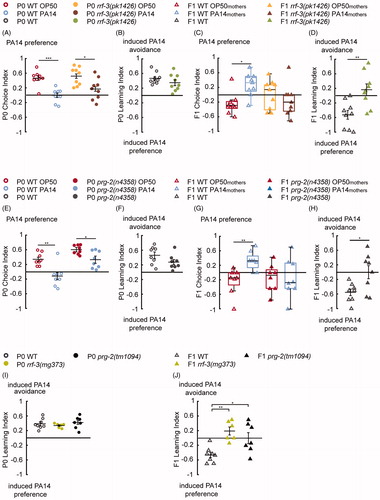
After sampling the olfactory preference in the naive and trained mothers, we harvested the embryos from the hermaphrodite mothers that remained on the control and training plates using a bleach solution, which dissolved P0 bodies and the associated bacteria and prevented the progeny from a direct contact with PA14. We cultivated F1s on OP50 under standard conditions and examined whether the learning experience of their P0 mothers modulated their olfactory behavior when they reached adulthood. To this end we tested F1 worms in the two-choice assay on plate and measured the difference between the choice index of the progeny of the naive mothers (F1 WT OP50mothers) and the choice index of the progeny of the trained mothers (F1 WT PA14mothers) (, Materials and methods). While the fast speed of the droplet assay allows us to sample learning in P0s before isolating F1 embryos carried by the trained hermaphrodites, the two-choice assay allows us to record and analyze chemotactic movements in F1s. Similar to the droplet assay, a positive Choice Index in the two-choice assay indicates a preference for PA14. In contrast with P0s, we found that the progeny of the trained mothers (F1 WT PA14mothers) showed a stronger preference for PA14 in comparison with the progeny of the naive mothers (F1 WT OP50mothers). This is reflected in a negative F1 Learning Index, which results from an average preference for PA14 in the progeny of the trained mothers and an average preference for OP50 in the progeny of the naive mothers (, data on the left side of the graphs depicted by empty triangles). Because the F1 progenies are isolated from trained hermaphrodites and cultivated on non-pathogenic OP50 without a direct exposure to PA14, this surprising result suggests that the food choices of the progeny are modulated by maternal exposure. A short exposure to the pathogen PA14 reduces the preference for the odorants of PA14 in mothers, but increases this preference in the progeny.
The endo-siRNA and piRNA pathways regulate parental experience modulation of behavior
We hypothesized that parental experience modulated the behaviour of the next generation through epigenetic mechanisms that regulate gene expression in progeny. In C. elegans, the endogenous RNA interference pathway can respond to environmental conditions to regulate physiological events in the offspring (Moore et al., Citation2019; Ni et al., Citation2016; Palominos et al., Citation2017; Posner et al., Citation2019; Rechavi et al., Citation2014; Schott, Yanai, & Hunter, Citation2014). Moreover, gene expression in the germline and oocytes is regulated by RRF-3, a RNA-dependent RNA polymerase that is needed for the biogenesis of endo-siRNAs (Gent et al., Citation2010; Han et al., Citation2009; Lee et al., Citation2006). The role of this pathway in olfactory behaviours has been previously demonstrated (Juang et al., Citation2013; Sims et al., Citation2016), which prompted us to ask how the disruption of the biogenesis of 26 G endo-siRNAs affected the modulation of olfactory choices in F1s. To this end we tested animals with a loss of function mutation in rrf-3(pk1426) (Simmer et al., Citation2002) and found that the mutant mothers displayed normal learning after 4-h training with PA14 (, data on the right side of the graphs depicted by filled circles). However, this learning experience did not alter the olfactory preference of their progeny, demonstrated by the similar choice indexes in rrf-3(pk1426) F1s from naive and trained mothers and a F1 Learning Index significantly different from that of wild type (WT) (, data on the right side of the graphs depicted by filled symbols). To further confirm the role of rrf-3, we tested another independently generated mutant allele, mg373, that harbours a missense mutation for a conserved catalytic residue (Pavelec, Lachowiec, Duchaine, Smith, & Kennedy, Citation2009). We found that rrf-3(mg373) mutant animals showed a similar phenotype to the rrf-3(pk1426) mutants in both P0s and F1s (; Supplemental Figure 2(A,B,D,E)). Together, these results indicate that the rrf-3-mediated endo-siRNA pathway regulates the modulation of offspring olfactory preference by parental experience.
We next examined the piRNA pathway that mainly regulates gene silencing and germline integrity (Ashe et al., Citation2012; Batista et al., Citation2008; Cox et al., Citation1998; Das et al., Citation2008; Lee et al., Citation2012; Weick & Miska, Citation2014). The piRNA population has been shown to be altered by exposure to environmental stressors, including pathogenic Pseudomonas aeruginosa strain PA14 (Belicard, Jareosettasin, & Sarkies, Citation2018). While the PIWI-clade Argonaute prg-1 has been previously implicated in regulating a form of pathogen avoidance that lasts for several generations (Moore et al., Citation2019), little is known about the function of its homolog prg-2 (Ashe et al., Citation2012; Batista et al., Citation2008; Cox et al., Citation1998; Das et al., Citation2008; Kasper, Gardner, & Reinke, Citation2014; Lee et al., Citation2012). We tested a deletion mutation, n4358, in the prg-2 gene. At least three alternatively spliced transcripts are predicted from the genomic locus of prg-2 and the deletion in n4358 affects all the transcripts. Two of the predicted transcripts encode pseudogenes and the third transcript encodes a homologous protein of PRG-1. We detected the third transcript in a wild-type cDNA library (Supplemental Figure 2(G,H)). PRG-1 regulates 21 U-RNAs (piRNAs) and mediates gene silencing, germline integrity and fertility (Ashe et al., Citation2012; Batista et al., Citation2008; Cox et al., Citation1998; Das et al., Citation2008; Lee et al., Citation2012; Weick & Miska, Citation2014). Although the putative PRG-2 protein is highly similar to PRG-1 and the prg-2 transcripts are found primarily in the germline (Hashimshony, Feder, Levin, Hall, & Yanai, Citation2015), the function of PRG-2 is not well characterized. We found that prg-2(n4358) P0s exhibited a significantly decreased preference for PA14 after training and generated learning indexes comparable to wild type (, data on the right side of the graphs depicted by filled circles). However, the F1s of the naive and trained prg-2(n4358) mothers displayed similar choice indexes for PA14, resulting in a F1 Learning Index significantly different from WT (, data on the right side of the graphs depicted by filled triangles). Furthermore, an independently generated deletion mutation, tm1094, in prg-2 similarly disrupted F1 Learning Index (, Supplemental Figure 2(C,F)). Taken together, these findings suggest the function of the piRNA pathway in transducing parental learning experience to the progeny and modulating offspring behavior.
Extending duration of parental exposure to PA14 switches attraction to aversion in progeny
We were intrigued by the increased olfactory preference for the pathogenic bacteria PA14 in the progeny of PA14-trained mothers. We asked whether stabilizing the parental environment by extending parental training with PA14 would generate a different response in the offspring. We found that training mothers with PA14 for 8 h also induced aversive learning of PA14 in the mothers (). However, the preference for PA14 in the progeny of the trained mothers was similar to that in the progeny of the naive mothers, which is reflected in a F1 Learning Index that has a mean value close to zero (). One feature in both the choice indexes and learning indexes of these progenies is their widespread distribution centred on 0. We noticed that the number of biological replicates in which the progeny of PA14-trained mothers preferred PA14 (negative Learning Index in F1) was similar to the number of biological replicates in which the progeny of trained mothers preferred OP50 (positive Learning Index in F1). This observation prompted us to examine whether there was a correlation between the learning of the mothers and the behavior of their progeny.
Figure 3. Extending parental training to 8 h switches offspring response from attraction to aversion. (A,B) Training with PA14 for 8 h decreases the preference for PA14 odorants in wild-type (WT) mothers (A), resulting in learned avoidance of PA14 (B), n = 12 biological replicates each, droplet assay. (C,D) The progeny of naive (F1 WT OP50mothers) and trained (F1 WT PA14mothers) mothers show similar preference for the pathogen, n = 12 biological replicates each, two-choice assay. (E) The Learning Index in F1s is linearly correlated with the Learning Index of their mothers after 8-h training. The progeny from the “WT P0 low learning index” group (light green stars) respond to PA14 differently from the progeny from the “WT P0 high learning index” group (dark grey stars). N = 12 biological replicates. (F) Schematics showing the trajectory of a F1 worm in the two-choice plate assay. (G–I) The movement speed (G), reversal rate (H) and the rate of omega bends (I) of F1s from mothers from the “WT P0 low learning index” group (n = 90 worms) and “WT P0 high learning index” group (n = 93 worms). (J) The effect of parental learning on body size of F1 WT OP50mothers (n = 90 worms) in comparison with F1 WT PA14mothers (n = 93 worms). (K) F1s of the trained mothers have similar body size. F1 WT PA14mothers low learning index (n = 45 worms) compared with F1 WT PA14mothers high learning index (n = 48 worms). (L) The F1 learning index is not correlated with the difference in F1 body size, n = 12 biological replicates. For all, ***p < 0.001, *p < 0.05. Data are normally distributed in A–E, G, J and presented with Mean ± SEM; data are not normally distributed in H, I, K and presented with box plots (showing median, first and third quartile, whiskers extending to values within 2.7 standard deviations). More details in statistical analyses are in Supplemental Table 1.
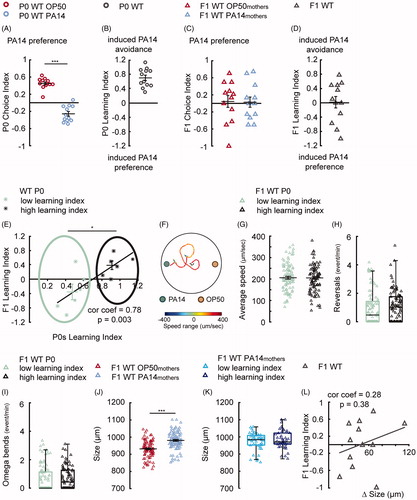
We plotted the Learning Index of F1 (y-axis) as a function of the Learning Index of their mothers in each independent experiment and found that they were linearly correlated—the more the mothers learned to avoid PA14, the more their offspring avoided the pathogen (). We calculated the average Learning Index of the P0s and divided the experiments in two groups—“P0 low learning index” and “P0 high learning index” (). Comparing these two groups we found that the F1s in experiments where mothers learned to avoid PA14 at a lower level, i.e. “P0 low learning index”, had a negative average Learning Index (). This indicates that in these experiments the progeny of trained mothers preferred PA14 more than the progeny of naive mothers, similarly to the progeny of 4-h trained mothers. In contrast, the F1s in the experiments where mothers learned to strongly avoid PA14, i.e. “P0 high learning index”, had a positive average F1 Learning Index (). This value suggests that the progeny of trained mothers avoided PA14 more than the progeny of naive mothers. Consistently, the F1s of the trained mothers from the “P0 high learning index” group avoided the pathogen significantly more than the F1s of the trained mothers from the “P0 low learning index” group (Supplemental Figure 3(A)). Together, these results show that the wide-spread behavioral responses to PA14 in the progeny of 8-h trained mothers result from the difference in the learning of their mothers.
We then asked how the progeny of the “P0 low learning index” group and the progeny of the “P0 high learning index” group differed in their chemotactic movements in the two-choice assay. We video tracked the worms and quantified several behavioral parameters in order to address if any specific features were modulated by parental experience and may underlie the different food odorant choices (, Material and methods). We found no difference in locomotor speed, reversal rate and frequency of omega bends (i.e. the big body bends that have the shape of the letter Omega) between the F1s from the “P0 low learning index” and the F1s from the “P0 high learning index” groups (). These results suggest that parental learning experience with PA14 regulates the choice of the offspring without affecting these specific locomotor parameters, and likely modulates offspring behavior by regulating a higher level of sensorimotor functions.
Meanwhile, we also noticed that F1s of the trained mothers were larger in body size than F1s of the naive mothers (). This intergenerational effect is consistent with previous findings showing that animals infected with PA14 retain their eggs more than worms raised on OP50 (Tan et al., Citation1999) and thus their progeny are kept inside the mother until a more advanced stage. Therefore, we examined if the difference in body size, which could be due to their difference in the developmental stage, could be related with the difference in the food choice. We found no differences when comparing the body size between F1s of the trained mothers in the “P0 low learning index” group and F1s of the trained mothers in the “P0 high learning index” group (), as well as no correlation between F1 Learning Index and the increase in body size in the progeny of the trained mothers compared with the progeny of the naive mothers (). In addition, we found that 8-h exposure to PA14, while inducing robust avoidance of PA14 in P0 mothers, did not alter the innate resistance to PA14 in F1s (Supplemental Figure 3(B)). Together, these results show that the body size and the innate immune resistance do not play a significant role in regulating parental experience-induced difference in offspring olfactory preference.
Next, we asked whether the learning experience in the P0 mothers trained with 8-h exposure to PA14 regulated F2s. We isolated F2 embryos from F1s and cultivated F2s under standard conditions on E. coli OP50. Similarly, we quantified the learning index in F2s in each experiment and found no correlation with the learning index of their F1 mothers (Supplemental Figure 4). These results indicate that the parental experience with PA14 only regulates the olfactory response of their first-generation progeny, potentially leaving the flexibility for F2s to respond to further changes in the environment.
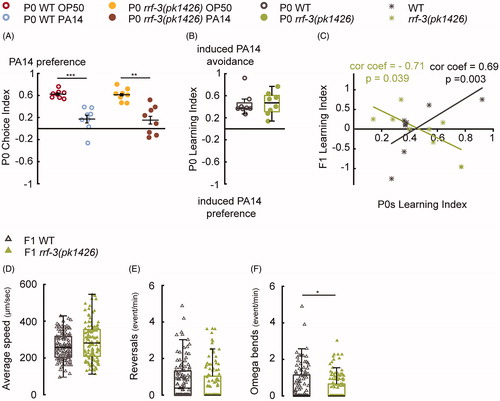
Furthermore, we found that mutating rrf-3 also disrupted the parental experience-induced behavioral change in the 8-h training experiments. Despite the wild-type learning ability in the rrf-3(pk1426) P0 mothers (), the learning index of rrf-3(pk1426) F1s is no longer correlated with the learning of their mothers (). Together, these results show that the rrf-3-mediated endo-siRNA pathway also regulates intergenerational effects generated by training P0 mothers for 8 h. In addition, we found that during the two-choice assay, while rrf-3 F1s had a locomotor speed and a rate of reversals comparable to those in wild-type F1s, their rate of omega bends was lower (). These results identify the regulation of omega bends as the potential downstream effector of the rrf-3-mediated pathway in regulating intergenerational effects.
Figure 4. The RRF-3-pathway regulates intergenerational effects generated by 8-h parental training with PA14. (A–C) The rrf-3(pk1426) mutant P0s display wild-type choice indexes (A) and learning (B) after 8-h training with PA14; however, the learning indexes in rrf-3(pk1426) F1s do not positively correlate with their mothers’ learning (C), n = 8 biological replicates for each genotype. P0s and F1s are respectively tested in droplet assay and two-choice assay. (D–F) The movement speed (D), reversal rate (E) and the rate of omega bends (F) in wild-type (WT) F1s (n = 107 worms) and rrf-3(pk1426) F1s (n = 115 worms). For all, ***p < 0.001, **p < 0.01, *p < 0.05. Data are normally distributed in A (Mean ± SEM) and not normally distributed in B, D–F (box plots showing median, first and third quartile, whiskers extending to values within 2.7 standard deviations). More details in statistical analyses are in Supplemental Table 1.
Discussion
Previous studies addressing the impact of parental experience on offspring show that intergenerational and transgenerational regulations are more complex than a unidirectional transmission of phenotypic traits acquired by the progenitors (Bohacek & Mansuy, Citation2015; Gapp et al., Citation2014; Palominos et al., Citation2017; Remy, Citation2010). Here, we find that a brief experience (4 h) with the pathogenic strain of Pseudomonas aeruginosa, PA14, while reducing the preference for the odorants of the pathogen in the trained mothers, increases the preference in the progeny. Furthermore, extending the maternal exposure time to 8 h switches the response of the progeny from attraction to avoidance in a way that is dependent on the experience of the mother. This intergenerational effect is limited to the first generation. Because the tested F1 progenies are isolated from the trained hermaphrodites at the end of the 4-h or 8-h training and because under standard conditions fertilized eggs spend 2–3 h to develop in utero before egg-laying (Hall, Herndon, & Altun, Citation2017), the behavioral effects in F1s that never directly interact with PA14 likely result from maternal exposure. Recently it has been shown that an even longer exposure to PA14 (24 h, starting at L4 larval stage) induces avoidance in mothers and their offspring for several generations through a separate mechanism involving a bacterial small RNA (Moore et al., Citation2019; Kaletsky, Moore, Parsons, & Murphy, Citation2019). Together, these results suggest that different maternal experience with the same type of bacteria can lead to diverse responses in progeny. They also suggest that the valence of the parental experience that is transmitted to the offspring is likely mediated by the physiological, but not behavioral, response of the parents.
After 4-h exposure to the pathogen, C. elegans adults shift their preference towards food sources that are non-pathogenic (Zhang et al., Citation2005), a behavioral defense against increased infection. However, when presented with the option between PA14 and no food, PA14-trained worms still prefer PA14 (Ha et al., Citation2010). In addition, in a lawn-leaving assay where worms feed on PA14 but can leave over time, almost all worms are still inside the lawn after 4 h and start to significantly leave only after 8 h [(Singh & Aballay, Citation2019) and unpublished results from our lab]. Under such conditions, it is conceivable that after a 4-h exposure PA14 is still perceived as a food source, albeit pathogenic, and positively modulates the preference of the progeny as the food consumed by their mothers. These findings are consistent with several studies in mice and humans showing that consuming foods containing certain taste or smell by mothers during gestation increases the preference of the progeny for the same taste or the odorant after birth (Hepper, Wells, Dornan, & Lynch, Citation2013; Todrank, Heth, & Restrepo, Citation2011). This modulation is likely to be adaptive since the food consumed by the mother is a potential source of safe food in the future. Extending the adult exposure to 8 h enhances the innate immune response to PA14 (Troemel et al., Citation2006), which likely represents a more stable presence of virulence, resulting in the switch of offspring behavior from attraction to avoidance. A longer exposure of 24 h to a pathogenic lawn grown under conditions that enhance bacterial pathogenicity (Moore et al., Citation2019) probably signals a constant presence of virulence that generates the transmission of aversive information for several generations. Together, these results on parental exposure to pathogenic bacteria are in agreement with observation in other species in which the intergenerational and transgenerational modulations of physiological and behavioral traits are influenced by the components of the diet of previous generations (Deas et al., Citation2019; Öst et al., Citation2014).
rrf-3 mutants are depleted in the 26 G RNA population, which affects endogenous gene expression during spermatogenesis, oogenesis and zygotic development (Gent et al., Citation2010; Han et al., Citation2009; Lee et al., Citation2006). Thus, our results suggest that these rrf-3-mediated gene expression programs can be modulated by parental experience in a way that affects the behavior of the progeny. Furthermore, one of the prg-2 transcripts encodes a homolog of a C. elegans Argonaute/Piwi-related protein PRG-1 that regulates piRNAs. Despite the high level of similarity between prg-1 and prg-2, the function of prg-2 is not well understood (Ashe et al., Citation2012; Batista et al., Citation2008; Cox et al., Citation1998; Das et al., Citation2008; Lee et al., Citation2012; Wang & Reinke, Citation2008). Our study reveals a role of prg-2 in regulating parental learning experience-induced modulation of olfactory preference. Thus, we show that both the endo-siRNA and piRNA pathways play a role in the modulation of olfactory preference in progeny after 4-h maternal exposure. Whether they affect the expression of the same genes is however an open question. Behaviorally, while both mutations abolish the increased preference for PA14 in F1s as a result of training their mothers for 4 h, their phenotypes are slightly different. This observation suggests that they may act in different ways to regulate parental experience-induced behavioral changes.
A major role of the endo-siRNA pathways is to modulate endogenous gene expression (Han et al., Citation2009; Lee et al., Citation2006; Mello & Conte, Citation2004; Okamura & Lai, Citation2008)—in its absence the basal level and the condition-dependent expression of these genes may be compromised. If parental experience-induced changes in the next generation depends on the modulation of gene expression in the progeny, the lack of the endo-siRNA pathway may compromise the effects of parental experience. Interestingly, a recent paper reports that the absence of a dsRNA-binding protein RDE-4 in neurons affects the pool of small RNAs in the germline in a heritable manner (Posner et al., Citation2019). Previous studies show that exogenously generated dsRNAs or bacterially derived small RNAs can also regulate gene expression or behavior in offspring and some of the processes affect the germ line (Ashe et al., Citation2012; Buckley et al., Citation2012; Fire et al., Citation1998; Kaletsky, Moore, Parsons, & Murphy, Citation2019; Palominos et al., Citation2017; Vastenhouw et al., Citation2006). Together, the results from these and our studies suggest that the normal function of the gene expression program regulated by small RNA pathways is important for parental experience-modulated behaviors. The findings showing that different parental experiences employ the small RNA pathways to generate modulatory effects in the offspring reveal the flexibility of the small RNA-mediated gene expression programs to encode various environmental conditions to regulate animal physiology across generations.
Author contributions
A.P., X.G., K.K. and Y.Z. designed the experiments, interpreted the results and wrote the manuscript. A.P., X.G. and K.K. performed the experiments and analyzed the data.
ineg_a_1819265_sm2899.pdf
Download PDF (682.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data and codes supporting this study are accessible in the Harvard Dataverse repository, https://doi.org/10.7910/DVN/UQAYAK
Additional information
Funding
References
- Ashe, A., Sapetschnig, A., Weick, E.M., Mitchell, J., Bagijn, M.P., Cording, A.C., … Miska, E.A. (2012). PiRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell, 150(1), 88–99. doi:10.1016/j.cell.2012.06.018
- Batista, P.J., Ruby, J.G., Claycomb, J.M., Chiang, R., Fahlgren, N., Kasschau, K.D., … Mello, C.C. (2008). PRG-1 and 21U-RNAs Interact to Form the piRNA Complex Required for Fertility in C. elegans. Molecular Cell, 31(1), 67–78. doi:10.1016/j.molcel.2008.06.002
- Belicard, T., Jareosettasin, P., & Sarkies, P. (2018). The piRNA pathway responds to environmental signals to establish intergenerational adaptation to stress. BMC Biology, 16(1), 1–14. doi:10.1186/s12915-018-0571-y
- Bohacek, J., & Mansuy, I.M. (2015). Molecular insights into transgenerational non-genetic inheritance of acquired behaviours. Nature Reviews. Genetics, 16(11), 641–652. doi:10.1038/nrg3964
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics, 77(1), 71–94.
- Buckley, B.A., Burkhart, K.B., Gu, S.G., Spracklin, G., Kershner, A., Fritz, H., … Kennedy, S. (2012). A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature, 489(7416), 447–451. doi:10.1038/nature11352
- Cox, D.N., Chao, A., Baker, J., Chang, L., Qiao, D., & Lin, H. (1998). A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes & Development, 12(23), 3715–3727. doi:10.1101/gad.12.23.3715
- Das, P.P., Bagijn, M.P., Goldstein, L.D., Woolford, J.R., Lehrbach, N.J., Sapetschnig, A., … Miska, E.A. (2008). Piwi and piRNAs Act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Molecular Cell, 31(1), 79–90. doi:10.1016/j.molcel.2008.06.003
- Deas, J.B., Blondel, L., & Extavour, C.G. (2019). Ancestral and offspring nutrition interact to affect life-history traits in Drosophila melanogaster. Proceedings of the Royal Society B: Biological Sciences, 286(1897), 20182778. doi:10.1098/rspb.2018.2778
- Dias, B.G., Maddox, S.A., Klengel, T., & Ressler, K.J. (2015). Epigenetic mechanisms underlying learning and the inheritance of learned behaviors. Trends in Neurosciences, 38(2), 96–107. doi:10.1016/j.tins.2014.12.003
- Dias, B.G., & Ressler, K.J. (2014). Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience, 17(1), 89–96. doi:10.1038/nn.3594
- Félix, M.A., & Duveau, F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biology, 10(1), 59. doi:10.1186/1741-7007-10-59
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., & Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391 (6669), 806–811. doi:10.1038/35888
- Gapp, K., Jawaid, A., Sarkies, P., Bohacek, J., Pelczar, P., Prados, J., … Mansuy, I.M. (2014). Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Neuroscience, 17(5), 667–669. doi:10.1038/nn.3695
- Garcia, J., Hankins, W.G., & Rusiniak, K.W. (1974). Behavioral regulation of the milieu interne in man and rat. Science (New York, N.Y.), 185(4154), 824–831. doi:10.1126/science.185.4154.824
- Gent, J.I., Lamm, A.T., Pavelec, D.M., Maniar, J.M., Parameswaran, P., Tao, L., … Fire, A.Z. (2010). Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans Soma. Molecular Cell, 37(5), 679–689. doi:10.1016/j.molcel.2010.01.012
- Ha, H. i., Hendricks, M., Shen, Y., Gabel, C.V., Fang-Yen, C., Qin, Y., … Zhang, Y. (2010). Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron, 68(6), 1173–1186. doi:10.1016/j.neuron.2010.11.025
- Han, T., Manoharan, A.P., Harkins, T.T., Bouffard, P., Fitzpatrick, C., Chu, D.S., … Kim, J.K. (2009). 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proceedings of the National Academy of Sciences USA, 106(44), 18674–18676. doi:10.1073/pnas.0906378106
- Hashimshony, T., Feder, M., Levin, M., Hall, B.K., & Yanai, I. (2015). Spatiotemporal transcriptomics reveals the evolutionary history of the endoderm germ layer. Nature, 519(7542), 219–222. doi:10.1038/nature13996
- Hepper, P.G., Wells, D.L., Dornan, J.C., & Lynch, C. (2013). Long-term flavor recognition in humans with prenatal garlic experience. Developmental Psychobiology, 55(5), 568–574. doi:10.1002/dev.21059
- Hall, D.H., Herndon, L.A., & Altun, Z. (2017). Introduction to C.elegans embryo anatomy. In Worm Atlas. doi:10.3908/wormatlas.4.1
- Horsthemke, B. (2018). A critical view on transgenerational epigenetic inheritance in humans. Nature Communications, 9(1):2973. doi:10.1038/s41467-018-05445-5
- Juang, B.-T., Gu, C., Starnes, L., Palladino, F., Goga, A., Kennedy, S., & L'Etoile, N.D. (2013). Endogenous nuclear RNAi mediates behavioral adaptation to odor. Cell, 154(5), 1010–1022. doi:10.1016/j.cell.2013.08.006
- Kaletsky, R., Moore, R.S., Parsons, L.L., & Murphy, C.T. (2019). Cross-kingdom recognition of bacterial small RNAs induces transgenerational pathogenic avoidance. biorXiv preprint. doi:10.1101/697888
- Kasper, D.M., Gardner, K.E., & Reinke, V. (2014). Homeland security in the C. elegans germ line: Insights into the biogenesis and function of pirnas. Epigenetics, 9(1), 62–74. doi:10.4161/epi.26647
- Kundakovic, M., Gudsnuk, K., Franks, B., Madrid, J., Miller, R.L., Perera, F.P., & Champagne, F.A. (2013). Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol a exposure. Proceedings of the National Academy of Sciences of the United States of America, 110(24), 9956–9961. doi:10.1073/pnas.1214056110
- Lee, H.C., Gu, W., Shirayama, M., Youngman, E., Conte, D., & Mello, C.C. (2012). C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell, 150(1), 78–87. doi:10.1016/j.cell.2012.06.016
- Lee, R.C., Hammell, C.M., & Ambros, V. (2006). Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA (New York, N.Y.), 12(4), 589–597. doi:10.1261/rna.2231506
- Liu, H., Yang, W., Wu, T., Duan, F., Soucy, E., Jin, X., & Zhang, Y. (2018). Cholinergic sensorimotor integration regulates olfactory steering. Neuron, 97(2), 390–405.e3. doi:10.1016/j.neuron.2017.12.003
- Mello, C.C., & Conte, D.J.. (2004). Revealing the world of RNA interference. Nature, 431(7006), 338–342. doi:10.1038/nature02872
- Miska, E.A., & Ferguson-Smith, A.C. (2016). Transgenerational inheritance: Models and mechanisms of non – DNA sequence – based inheritance. Science (New York, N.Y.), 354(6308), 59–782. doi:10.1126/science.aaf4945
- Moore, R.S., Kaletsky, R., & Murphy, C.T. (2019). Piwi/PRG-1 argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell, 177(7), 1827–1841. doi:10.1016/j.cell.2019.05.024
- Ni, J.Z., Kalinava, N., Chen, E., Huang, A., Trinh, T., & Gu, S.G. (2016). A transgenerational role of the germline nuclear RNAi pathway in repressing heat stress-induced transcriptional activation in C. elegans. Epigenetics & Chromatin, 9(1), 3–15. doi:10.1186/s13072-016-0052-x
- Okamura, K., & Lai, E.C. (2008). Endogenous small interfering RNAs in animals. Nature Reviews Molecular Cell Biology, 9(9), 673–678. doi:10.1038/nrm2479
- Öst, A., Lempradl, A., Casas, E., Weigert, M., Tiko, T., Deniz, M., … Pospisilik, J.A. (2014). Paternal diet defines offspring chromatin state and intergenerational obesity. Cell, 159(6), 1352–1364. doi:10.1016/j.cell.2014.11.005
- Palominos, M.F., Verdugo, L., Gabaldon, C., Pollak, B., Ortíz-Severín, J., Varas, M.A., … Calixto, A. (2017). Transgenerational diapause as an avoidance strategy against bacterial pathogens in Caenorhabditis elegans. mBio, 8(5), 1–18. doi:10.1128/mBio.01234-17
- Pavelec, D.M., Lachowiec, J., Duchaine, T.F., Smith, H.E., & Kennedy, S. (2009). Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics, 183(4), 1283–1295. doi:10.1534/genetics.109.108134
- Posner, R., Toker, I.A., Antonova, O., Star, E., Anava, S., Azmon, E., … Rechavi, O. (2019). Neuronal small RNAs control behavior transgenerationally. Cell, 177(7), 1814–1826.e15. doi:10.1016/j.cell.2019.04.029
- Rechavi, O., Houri-Ze'evi, L., Anava, S., Goh, W.S.S., Kerk, S.Y., Hannon, G.J., & Hobert, O. (2014). Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell, 158(2), 277–287. doi:10.1016/j.cell.2014.06.020
- Remy, J. (2010). Stable inheritance of an acquired behavior in Caenorhabditis elegans. Current Biology : CB, 20(20), R877–R875. https://doi.org/10.1016/j.cub.2010.08.013.Acknowledgements. doi:10.1016/j.cub.2010.08.013
- Samuel, B.S., Rowedder, H., Braendle, C., Félix, M.A., & Ruvkun, G. (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proceedings of the National Academy of Sciences of the United States of America, 113(27), E3941–E3949. doi:10.1073/pnas.1607183113
- Schott, D., Yanai, I., & Hunter, C.P. (2014). Natural RNA interference directs a heritable response to the environment. Scientific Reports, 4, 7387. doi:10.1038/srep07387
- Simmer, F., Tijsterman, M., Parrish, S., Koushika, S.P., Nonet, M.L., Fire, A., … Plasterk, R.H.A. (2002). Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. Elegans hypersensitive to RNAi. Current Biology : CB, 12(15), 1317–1319. doi:10.1016/S0960-9822(02)01041-2
- Sims, J.R., Ow, M.C., Nishiguchi, M.A., Kim, K., Sengupta, P., & Hall, S.E. (2016). Developmental programming modulates olfactory behavior in C. elegans via endogenous RNAi pathways. eLife, 5, 1–26. doi:10.7554/eLife.11642
- Singh, J., & Aballay, A. (2019). Microbial colonization activates an immune fight-and-flight response via neuroendocrine signaling. Developmental Cell, 49(1), 89–99.e4. doi:10.1016/j.devcel.2019.02.001
- Tan, M.W., Mahajan-Miklos, S., & Ausubel, F.M. (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America, 96(2), 715–720. doi:10.1073/pnas.96.2.715
- Todrank, J., Heth, G., & Restrepo, D. (2011). Effects of in utero odorant exposure on neuroanatomical development of the olfactory bulb and odour preferences. Proceedings. Biological Sciences, 278(1714), 1949–1955. doi:10.1098/rspb.2010.2314
- Troemel, E.R., Chu, S.W., Reinke, V., Lee, S.S., Ausubel, F.M., & Kim, D.H. (2006). p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genetics, 2(11), e183. doi:10.1371/journal.pgen.0020183
- Vastenhouw, N., Brunschwig, K., Okihara, K.L., Muller, F., Tijsterman, M., & Plasterk, R.H.A. (2006). Long-term gene silencing by RNAi. Nature, 442(7105), 882. doi:10.1038/442882a
- Wang, G., & Reinke, V. (2008). A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Current Biology : CB, 18(12), 861–867. doi:10.1016/j.cub.2008.05.009
- Weick, E.M., & Miska, E.A. (2014). piRNAs: From biogenesis to function. Development (Cambridge, England), 141(18), 3458–3471. doi:10.1242/dev.094037
- Weigel, D., & Colot, V. (2012). Epialleles in plant evolution. Genome Biology, 13(10), 249–246. doi:10.1186/gb-2012-13-10-249
- Zhang, Y., Lu, H., & Bargmann, C.I. (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature, 438(7065), 179–184. doi:10.1038/nature04216
