Abstract
Purpose: To elucidate the thermal impact on biological processes the first three hours after hyperthermia (HT).
Materials and methods: Tumor samples from BT4An rat tumors were transplanted subcutaneously to the right hind foot of BD IX rats, treated with local water-bath HT (mean tumor temperature of 43°C) for one hour, and analyzed for changes in global gene expression the first three hours after treatment with HT. Samples from a corresponding in vitro experiment were also analyzed. Differentially expressed genes in vivo were mapped to a gene ontology (GO) database in search for biological processes overrepresented after treatment with HT. Selected genes were verified using Taqman quantitative RT-PCR.
Results: 1213 genes were differentially expressed after HT treatment in vivo. Processes overrepresented by GO mapping could be sorted into 10 main functional groups; apoptosis, transcription, immune system, blood vessels, metabolism/protein modifications, differentiation, cell signaling, response to stimulus, transport and cytoskeleton. We detected reduced mRNA levels of a wide range of T-cell, natural killer cell and antigen presenting cell-related genes after HT.
Conclusion: To our best knowledge, this is the first global microarray analysis of a malignant tumor treated with HT in vivo. We highlight biological processes affected by HT. RNA expression profiles also indicate a rapid suppression of the immune system within tumors by local HT.
Introduction
Hyperthermia (HT), i.e. elevation of temperature above 41°C, has for a long time been investigated for the treatment of solid neoplasms. HT has limited anti-tumor effect alone, but potentiates, by more than an additive effect, radiation and many chemotherapeutics Citation[1], Citation[2]. HT may also be used to enhance liposomal drug delivery to tumors Citation[3], especially when administered concomitantly Citation[4]. HT inhibits the endothelial cell proliferation (angiogenesis) in vitro and in vivo, in a dose dependent manner Citation[5], destroys tumor blood vessels (angiotoxic effect) and may reduce tumor blood perfusion Citation[5–8]. The cell membrane, cytoskeleton, cytoplasm and nucleus are targets for hyperthermic therapy Citation[9–12]. HT interacts with the immune defense Citation[13] and interferes with mitosis Citation[9], Citation[12]. Protein denaturation and aggregation, proposed to be important for cytotoxicity, heat radiosensitization as well as thermotolerance, have also been reported Citation[10]. Heat shock proteins (HSPs) play an important part in development of thermotolerance Citation[10], Citation[14], but they also have a role in innate and adaptive immunity Citation[13], Citation[15–17]. HT is a strong challenge for the tumor cells, which may respond dually by signaling towards both survival and cell death Citation[10]. The sum of all these signals determines the fate of the cell.
Realizing the power of HT combined with chemotherapeutics, there is a need for better understanding of the molecular basis for the effects of HT. Several biological processes are affected by hyperthermia, but the genes involved have not been thoroughly investigated. To address this issue, we therefore performed the first global scale cDNA microarray screen of a malignant tumor treated with HT in vivo versus sham treatment. The data is expanded by corresponding experiments in vitro. Analyses of early gene expression changes within three hours after end of treatment day zero were performed on fresh frozen tissue samples from a collection of tumors harvested in a previous study. In this study, HT alone or combined with metronomic cyclophosphamide (CTX), significantly delayed tumor growth of a glioblastoma-like tumor model in rats Citation[18]. Tumor cells were also exposed to HT in culture, and cells harvested for gene expression analyses. In the in vivo material, we focused on genes differentially expressed after treatment with HT. These genes were sorted for association with biological processes, and selected overrepresented processes are discussed. Differentially expressed genes in vivo were compared with corresponding genes in vitro to highlight whether the intact tumor cell microenvironment influenced expression of the genes involved. Selected findings were qualitatively verified by quantitative RT-PCR. Immunohistochemistry of CD8 and MHC Class II Ia was performed to verify infiltration of immune cells in the tumors.
Materials and methods
Tumor tissue
Tissue was taken from tumors treated as previously described Citation[18]. In short, we used BDIX/HanFoss rats bearing a BT4An tumor (an aggressive glioma) subcutaneously on the hind leg. Animals were allocated to the following groups: HT at 44.1°± 0.1°C water-bath for one hour (HT) (yields mean tumor temperature of 43°C), CTX 35 mg/kg intraperitoneally administered as repeated doses 3 times a week for a total of 6 treatments (CTX), combined therapy with HT immediately following the first injection of CTX (day zero) (CTX-HT), or controls injected with saline. Samples were collected at 0, 45, 90 and 180 min after end of treatment. The animals had then received the HT treatment and/or a single low dose of CTX or saline. The rats were killed using CO2 gas, tumor harvested and frozen directly on liquid nitrogen.
Cell line
To establish a cell line, the BT4An tumor was trypsinized and the cells were plated in 25 cm2 culture bottles (Nunc Inc.) with 10 ml Dulbecco's minimal essential medium (D-MEM, + 4500 mg/l glucose, + GlutaMax, + Pyruvate) (Gibco) with 10% heat inactivated fetal calf serum. Non-adherent cells were washed off the following day and the cells left to grow until confluence. To remove rat fibroblasts, cells were trypsinized and plated on Soft-Agar, 0.5% agarose in D-MEM, for two weeks.
In vitro hyperthermia
We used 20 x 106 cells, and changed to conditioned medium two hours before the experiment. The cells were dissociated from the bottle using a cell scraper and centrifuged at 1100 rpm for 3 min. The cells were then resuspended in 24 ml of the same medium, and 0.5 ml was transferred to each of 44 sample tubes (1.5 ml Eppendorf tubes). We randomly selected 24 of these as controls for further culture at 37°C. Twenty samples received HT, precisely controlled in a water-bath at 43.0°+/− 0.1°C for 60 min. Following treatment, the samples were further cultivated at 37°C, similar to the controls. Four control samples were harvested at start of the experiment (−60 min). Next, four samples were harvested from each of the two groups at the following time points after initiation of hyperthermia: −30, 0 (end of treatment), 60, 180 and 300 min (). Each sample was homogenized before 250 µl was centrifuged for 30 s and the pellet dissolved in 350 µl lysisbuffer (RNeasy Mini Kit, Qiagen, Valencia, CA, USA) for extraction of RNA.
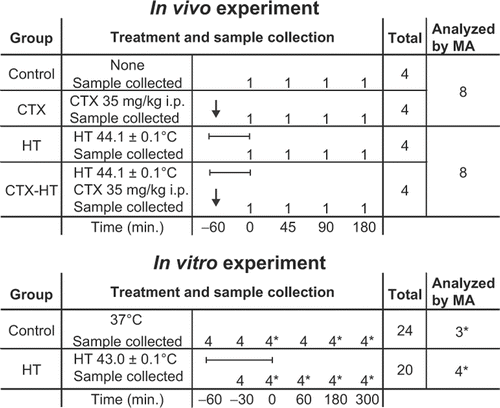
Figure 1. Outline of sample collection. We analyzed global gene expression at 0, 45, 90 and 180 min in the four treatment groups in vivo, at 0, 180 and 300 min in the in vitro control group (3 samples), and at 0, 60, 180 and 300 minutes in the in vitro HT group (4 samples). *, equal amounts of total RNA from four parallel samples were mixed and analyzed as one by the microarrays; bar, indicate HT treatment session; arrow, CTX or saline injected at start of HT; MA, microarray.
DNA sequencing
PCR primers were either chosen based on previous information (p53 Citation[19] and CDKN2A Citation[20], Citation[21]) or designed (). RT-PCR was performed using 10 x PCR buffer II, Amplitaq Gold Polymerase, ddH2O, MgCl2 and dNTP mix. In some of the reactions betain was added (all reagents from Applied Biosystems). 35 RT-PCR cycles were performed on a thermal cycler using the following programs: 14 min denaturation (denat.) at 94°C; for p53, CDKN2A and Hras 35 cycles of (20 s denat. at 94°C, 20 s annealing (anneal.) at X°C, 30 s elongation (elong.) at 72°C) and for Nras, Kras2 and Braf 35 cycles of (30 s denat. at 94°C, 30 s anneal. at X°C, 90 s elong. at 72°C); then 7 min elong. at 72°C and hold at 4°C. CDKN2A, Hras, Nras and KRAS2 were annealed at 58°C, p53 at 55°C and Braf at 61°C. The products were separated on a 1% agarose gel. Excess primers and nucleotides were removed by adding 1 µl Exosap (Applied Biosystems) and incubated at 37°C for 15 min and then 80°C for 15 min. DNA sequencing reactions (program: 20 s denat. at 96°C; 25 cycles of (10 s denat. at 96°C, 5 s anneal. at X°C, 4 min elong. at 60°C); then hold at 4°C) were performed on ABI 3100 or 3730 genetic analyzers (Applied Biosystems) using BigDye version 3.1 (Applied Biosystems) following standard protocols. Sequencing clean-up was performed using Millipore Multiscreen 384-SEQ plates as recommended by the manufacturer.
Table I. RT-PCR primer sequences
RNA purification and cDNA synthesis
Total RNA from tumor tissue and cell culture was extracted using protocol ‘Animal Tissue’ and ‘Animal Cells I’, respectively, from the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Total RNA concentrations and quality of both the in vivo and the in vitro samples were assessed using Nanodrop Spectrophotometer (Nanodrop Technologies) and all samples (total RNA) had OD260/280 in the range of 2.04–2.13. Samples to be used for real time RT-PCR were DNase I treated (Ambion), and cDNA synthesis was performed according to standard protocols Citation[18]. The inputs were 80 ng DNase I treated RNA from the tissue samples and 60 ng DNase I treated RNA from the cell culture samples, and final volumes of the cDNA reactions were 50 µl. Quality of samples (cRNA) to be used for microarrays was assessed with Nanodrop Spectrophotometer and OD260/280 were all in the range of 2.19–2.28. Total RNA from the tissue samples were also assessed with Agilent Technologies 2100 Bioanalyzer.
Microarray experiment
For analyses of global gene expression we used Applied Biosystems Rat Genome Survey Microarray analyzing 26.857 rat genes. In vitro we had four parallel samples at each time point. Equal amounts of total RNA from the four parallels were mixed and analyzed as one sample on the microarrays. Two µg total RNA was used as input in the cDNA reaction, using Applied Biosystems Chemiluminescence RT-IVT Labeling Kit (Part no. 4346877 Rev D). Gene expression was detected using Applied Biosystems Chemiluminescence Detection Kit (Part no. 4346875 D) and an Applied Biosystems 1700 Chemiluminiscent Analyzer. Files were imported as single channel data and analyzed with J-Express Pro 2.7 (Molmine AS, Bergen, Norway). The Gene Ontology Mapping Module sorts genes into three main groups: molecular function, cellular component and biological process, based on a database from the Gene Ontology Consortium Citation[22]. Within each main group, there are several thousand subgroups (GO terms). To use this module we imported a GO OBO v1.2 file and a gene_association RGD file of rats, on the 20th February 2007 from the Gene Ontology Consortium homepage: www.geneontology.org. The Gene Ontology Mapping module reported GO terms with a relative over-representation of genes in the subset of differentially expressed genes compared to all detected genes.
Real time RT-PCR
Real time RT-PCR was conducted as described previously Citation[18]. The samples were analyzed using the standard curve method Citation[23]. All samples were expressed as ratios of relative amount of probe of interest versus an internal control probe. As internal control, we used ß-actin (NM_031144) (Applied Biosystems Assay ID Rn00667869_m1) for analyses of the tissue samples and acidic ribosomal phosphoprotein P0 (ARBP) (NM_022402) (5′ ctggctcccaccttgtctccagtctttatc 3′ (primer), 5′cattgaaatcctgagcgatgtg 3′ (sense), 5′agatgttcaacatgttcagcggtgt 3′ (antisense), (Custom designed OliGold oligonucleotides from Eurogentec, Belgium) for analyses of cell samples. We investigated gene expression of early growth response 1 (EGR1) (NM_012551) (Rn00561138), jun oncogene (JUN) (NM_021835) (Rn00572991_s1), activating transcription factor 3 (ATF3) (NM_012912) (Rn00563784) and FBJ murine osteosarcoma viral oncogene homolog (FOS) (NM_012954) (Rn00564121_m1). (All probes except ARBP from Applied Biosystems.)
Immunohistochemical staining of CD8 and MHC Class II Ia
Immunohistochemical staining of Major Histocompatibility Complex (MHC) Class II Ia (ab6403, Abcam Inc., UK) and CD8 alpha (MCA48G, AbD Serotec, Oxford, UK) antigen was performed on formalin fixed and paraffin embedded tissue, incubated at 60°C for one hour and rehydrated with xylene and ethanol in decreasing concentrations. Target retrieval was achieved using a heat mediated pressure cooker method with 0.05% Citraconic anhydride buffer, pH 7.4 (Fluka, Sigma-Aldrich, Switzerland) for MHC Class II. CD8 staining did not require target retrieval. The slides were incubated one hour with primary antibody diluted 1:50 (MHC Class II Ia) and 1:400 (CD8) in Antibody Diluent (cat. no. S0809). Endogenous peroxidase was blocked using 3% hydrogen peroxide and detection was performed using biotin-linked LSAB2® System-HRP for use on rat specimens (cat. no. K0609). The slides were further processed by incubation with 3,3′-diaminobenzidine (DAB, from LSAB® System-HRP kit, cat. no. K0673), and after rinsing, counterstained with Hematoxylin (cat. no. S2020). They were then dehydrated using increasing concentrations of ethanol and xylene, and mounted using Eukitt quick-hardening mounting medium (cat. no. 03989, Fluka, Sigma-Aldrich, Switzerland). (All reagents from Dako, Denmark, when not otherwise specified.)
The microscopy slides were scored by two investigators blinded for the treatment groups, and counted in ten visual fields (x400) per slide, five from the periphery and five from the center of each tumor. Some tumors had a rim of mononuclear cells infiltrating the periphery of the tumor. Average number of cells per x400 field in each tumor was counted, and the average percentages of CD8 positive cells (cytotoxic T-cells and NK cells) and MHC Class II Ia positive cells (B-lymphocytes, various epithelial cells, dendritic cells and macrophages) per slide were estimated ().
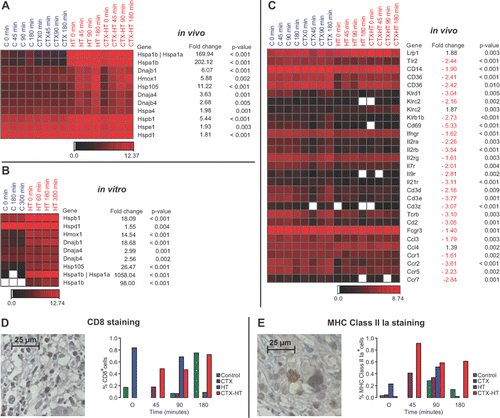
Figure 2. Supervised screen for HSP and genes involved in the immune system in the dataset of 1213 genes found differentially expressed after HT by SAM analysis in vivo. (A) HSPs differentially expressed in vivo and (B) the corresponding HSP genes in vitro. (C) genes involved in the immune system, especially HSP, T-cell, NK cell and APC related genes. Only a few genes involved in this immune system cluster were also differentially expressed in vitro (data not shown).Genes analyzed and displayed as single channel data, p-values, Student's t-test. (D) and (E) Immunohistochemical staining of CD8 positive cells (cytotoxic T-lymphocytes and NK cells) (D) and MHC class II Ia positive cells (B-lymphocytes, dendritic cells and macrophages) (E), pictures display representative staining from the periphery of a tumor treated with hyperthermia, percentage of positive cells displayed in the graphs. Pictures taken at x400 resolution, 25 µm scale bar. The figures have been proportionally adjusted in size after composition. No other adjustments of pictures.
Ethics
All the experiments and procedures described regarding the treatment of animals were approved by the local responsible laboratory animal science specialist under the surveillance of the Norwegian animal research authority. The experiments were conducted in accordance with the laws and regulations controlling experiments on live animals in Norway.
Statistics
Statistical analysis of microarrays (SAM) Citation[24] was conducted using J-Express pro 2.7 software package (Molmine AS, Bergen, Norway). The SAM analysis ranked the genes after a false discovery rate (FDR) indicating the percentage of genes in the list differentially expressed by chance. Since the analysis incorporates the probability of random findings in a large dataset, the FDR value is higher than the p-value for an individual gene. The FDR threshold was set at 2.5%. Genes found to be differentially expressed were also analyzed with a Student's t-test (feature subset selection (FSS) in J-Express) to calculate the p-value of individual genes. For statistical analysis of changes in single gene expression, we refer to the p-value of the Student's t-test. Over-representation of gene clusters, mapped to GO terms, was analyzed by the Gene Ontology Mapping module (J-Express) using a Student's t-test. In vitro, four samples from tumors treated with HT and three from controls () were analyzed. The number was too small for a formal SAM analysis, and we therefore used the Student's t-test (FSS) for the analysis of difference, setting the significance level at alpha = 0.05.
Real time RT-PCR and IHC results were analyzed with a Student's t-test to verify the hypothesis that there is a difference between the two groups (HT versus control) (α = 0.05). Real time RT-PCR and IHC statistics were performed using SPSS 14.02 statistical package (SPSS Inc.).
Results
Mutational status and characteristics of the tumor
Tumor protein p53, cyclin-dependent kinase inhibitor 2A (CDKN2A) (encoding p16INK4 and p14ARF), v-raf murine sarcoma viral oncogene homolog B1 (BRAF), v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS2), neuroblastoma ras oncogene (NRAS) and Harvey rat sarcoma viral (v-Ha-ras) oncogene homolog (HRAS) genes were sequenced to characterize the mutational status of the tumor. DNA sequencing of RT-PCR products of p53, CDKN2A, BRAF, KRAS2, NRAS and HRAS genes, revealed an activating heterozygote mutation in BRAF (V600E). No other mutations were detected in the six genes investigated.
Microarray analyses
A total of 23 microarray analyses were performed, 16 from the in vivo experiment (one sample of the C, CTX, HT and CTX-HT treatment groups for each of the four time-points), and 7 from the in vitro experiment (one sample from four time-points from the HT group and one sample from three time points from the C group) ().
In vivo, mRNA expression of 21,385 genes was detected by the microarrays. There were marked differences in mRNA expression of the samples treated with HT or CTX-HT versus those treated with CTX or saline. Only subtle differences in gene expression were seen between tumors treated with CTX and saline, and between HT treated tumors with or without CTX. To find genes differentially expressed due to the HT treatment in vivo, the samples were therefore sorted in two groups, one with samples from tumors treated with HT alone or the combination of HT and CTX, the other group with samples from tumors treated with CTX alone or saline (). We refer to this grouping (HT/CTX-HT versus saline/CTX) when we discuss HT-induced changes in gene expression. A formal SAM analysis of the samples treated with HT versus those treated without HT, revealed 1,213 differentially expressed genes. Individual p-values for single genes in this group varied from <0.001 to 0.014. The 1,213 genes were imported into the Gene Ontology Mapping Module. We focused on biological processes, and identified a total of 120 GO terms that were overrepresented in this subset of genes. Some of these terms were subgroups of others identifying the same genes. The overrepresented GO terms were classified in ten groups; apoptosis, transcription, immune system, blood vessels, metabolism/protein modifications, differentiation, cell signaling, response to stimulus, transport and cytoskeleton (). Selected processes are highlighted in . Genes in the immune system cluster displayed quite uniform mRNA expressions. Of the 50 different genes detected by the unsupervised analysis, 80% had reduced and 20% had increased mRNA levels after HT.
Table II. GO biological processes overrepresented (p < 0.05) in a cluster of 1213 genes differently expressed (SAM analysis) after hyperthermia in a rat glioblastoma like tumor model.
In vitro, mRNA expression of 26,038 genes was detected by the microarrays. The Student's t-test disclosed 1,620 genes that were differentially expressed after treatment with HT in vitro. Of the 1,213 genes found in vivo, 127 were also found in vitro. The majority of genes involved in the large ‘immune system’ group in vivo were not found differentially expressed in vitro.
Table III. Selected biological processes representing five main groups of GO terms overrepresented after HT.
From a supervised analysis of the 1,213 differentially expressed genes in vivo, we identified a number of HSPs. All of these had a marked increase in mRNA expression, both in vivo and in vitro (). Clustered with the HSPs, we also identified DNAJB4 mRNA 2.7 times up regulated in vivo and 2.6 times up regulated in vitro. Several genes involved in the interplay between natural killer (NK) cells, antigen presenting cells (APC), cytotoxic T-lymphocytes and HSP were also found to be differentially expressed in HT/CTX-HT samples (). We detected four receptors for HSP on APC cells Citation[17]; low density lipoprotein receptor-related protein 1 (Lrp1), toll-like receptor 2 (Tlr2), CD14 antigen (CD14) and CD36 antigen (CD36). We registered increased mRNA levels of Lrp1 and decreased mRNA levels of Tlr2, CD14 and CD36 after HT. Three important NK cell receptors Citation[15], killer cell lectin-like receptor, subfamily D, member 1 (Klrd1) alias CD94, killer cell lectin-like receptor subfamily B member 1B (Klrb1b) alias CD161, and CD69 antigen all had reduced mRNA levels after HT. We also detected reduced mRNA levels of IFNγ receptor, IL2 receptor alpha chain (IL2Rα) and beta chain (IL2Rß), the important common γ subunit (IL2Rγ), IL7R, IL9R and IL21R. Important members of the CD3 complex found in cytotoxic T-lymphocytes also displayed reduced mRNA levels after HT, including CD3 antigen, δ polypeptide (CD3δ) and ϵ polypeptide (CD3ϵ). Furthermore, we also detected some chemokines and chemokine receptors differentially expressed after treatment with HT; Chemokine, CC motif, Ligand 3 and 4 (CCL3 and CCL4) and the Chemokine, CC motif, Receptor 1, 2, 5, 7 (CCR1, CCR2, CCR5, CCR7). All except CCL4 had reduced mRNA levels after HT. Reduced mRNA levels after HT of the T-cell receptor ß chain (TCRß), and CD2 antigen (receptor on T-cells and some NK cells) and Fc receptor, IgG, low affinity III (Fcgr3) (receptor on NK cells) were also registered.
Unsupervised analysis (GO mapping) revealed up regulation of a range of dual specificity phosphatases (DUSPs) among the 1,213 differentially expressed genes, and we sorted them into the protein amino acid dephosphorylation cluster (). An additional DUSP gene was found by supervised analysis of the 1,213 genes. This gene was DUSP1 which was induced 10- fold in vivo and 17-fold in vitro.
Real time – RT-PCR
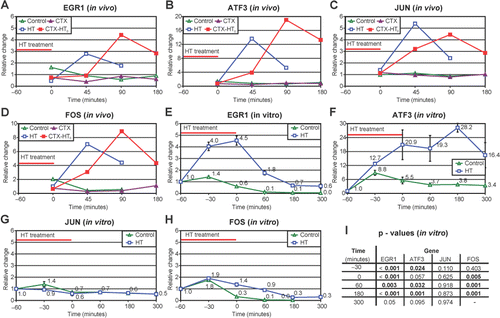
Real time quantitative RT-PCR confirmed a marked increase in gene expression of EGR1, ATF3, JUN and FOS in vivo (). In vitro, quantitative RT-PCR confirmed a marked increase in expression of EGR1 and ATF3 and a minor increase in expression of FOS. There was no increase in expression of JUN in vitro ().
Figure 3. Real time RT-PCR results in vivo (A–D) and in vitro (E–H). All samples are expressed as ratios of concentration of probe of interest versus a control probe, adjusted so that the mean of the controls at the first time point (in vitro) or all time points (in vivo) equals one. (I) statistical results in vitro, p-values from Student's t-test are presented.
Immunohistochemical staining
CD8 positive cells had in general strong staining of the cell membrane and displayed clear lymphocyte morphology. These cells represent cytotoxic T-cells and NK cells and were generally found at the periphery of the tumors. We detected variable degrees of CD8 positive cell infiltration in the sham tumors and tumors treated with CTX, median 0.07% (range: 0.00–0.75%). A higher amount of CD8 positive cells were detected in the peripheral rim of tumors treated with HT/CTX-HT, but the overall median number of CD8 positive cells in these tumors, 0.48% CD8 positive cells (range: 0.00–0.84%), was not significantly different from controls and CTX treated tumors ( D). A median of 0.13% MHC class II Ia positive cells (B-cells, various epithelial cells, dendritic cells and macrophages) were detected in controls and CTX treated tumors (Range: 0.02–0.34%). In accordance with the staining of CD8, there was a higher amount of MHC class II Ia positive cells in the periphery of the tumors treated with HT/CTX-HT, but the overall median of 0.55% positive cells (Range 0.02–0.91) was not significantly increased compared to controls and CTX treated tumors ().
Discussion
We designed this study to elucidate early molecular mechanisms responsible for the effects of HT. In general, treatment of an aggressive tumor will always induce a stress response to facilitate survival of the cells. However, the goal of cancer therapy is to overcome survival mechanisms by growth inhibitory and death signals. In previous work Citation[18], we have shown a growth inhibitory effect of HT, especially when combined with two weeks’ treatment with low doses of CTX. TUNEL analysis disclosed apoptosis as early as 90 min after treatment with HT, indicating that growth inhibitory and apoptotic signals were stronger than those of survival, and that differentially regulated genes in the first hours after treatment may have been involved in directing to cell death.
Mutational status
Sequence analyses of six genes often mutated in malignancies revealed the most common activating mutation of BRAF (V600E) Citation[25]. Mutations in genes may affect biological processes such as cell survival, oncogenic and apoptotic signaling. The activating BRAF mutation indicates a constitutive activation of the Mitogen activated protein kinase (MAPK) signaling pathway. This regulates gene expression and is involved in a wide range of biological processes inside the cell. Wild-type (wt) p53 is important for induction of apoptosis, as will be discussed in more detail later.
Gene expression
After treatment with HT, 1,213 genes in vivo and 1,620 genes in vitro were differentially expressed. The Student's t-test used to analyze the in vitro results does not incorporate the statistical uncertainty of large datasets. To minimize the risk of false positive findings, we therefore analyzed the in vitro results in light of the in vivo results and found 127 genes that were differentially expressed both in vivo and in vitro. Many responses inside the cells are dependent on cell to cell communication and signaling in the microenvironment that are lost in vitro. This may be a reason for retrieving as few as 127 genes in vitro, and highlights the importance of in vivo assays.
Validity
To confirm external validity, we compared our data with the results from a study by Kato et al. Citation[26]. They performed gene expression analyses (Agilent Human 1 cDNA microarray analyzing 12,814 genes) of HeLa cells cultivated for up to six hours after treatment with HT at 44.0°C for one hour in vitro Citation[26]. In HeLa cells, 664 genes were differentially expressed after HT, and the authors focused on DNAJ (Hsp40) homolog, subfamily B, member 4 (DNAJB4), heat-shock 70-kD protein (Hsp70), heat-shock 90-kD protein (Hsp90, HSPC), growth arrest and DNA damage-inducible protein 45 (GADD45), EGR1, JUN and FOS which all displayed increased mRNA levels after treatment with HT. We had a similar discovery rate in our material, revealing 1,213 differentially expressed genes (analyzing 26,857 genes) and largely our data comply with these previously published results Citation[26]. We detected increased gene expression of a range of HSPs () both in vivo and in vitro, among them DNAJB4 and HSPA1B (Hsp70). Gene expression of both GADD45G and GADD45B was induced in vivo, but only GADD45B was induced in vitro. Elevated mRNA levels of EGR1 were detected after HT in vivo and in vitro. FOS and JUN exhibited elevated mRNA levels in vivo but not in vitro. The expression profiles of EGR1, FOS and JUN were analyzed with real time RT-PCR to support the internal validity of the microarray analysis. Real time RT-PCR confirmed the results of the microarrays, with the exception of FOS.
Biological processes overrepresented after treatment with hyperthermia
Apoptosis
GO mapping revealed three large clusters involved in apoptosis overrepresented in the subgroup of differentially expressed genes, indicating that apoptotic mechanisms are affected by the therapy. However, these genes displayed mixed alterations in gene expression, and therefore we cannot make a conclusion about the impact on the cells based on these data alone. We previously reported TUNEL (terminal deoxynucleotidyl biotin – dUTP nick-end labeling) analysis which clearly indicated high rate of apoptosis as early as 90 minutes following end of treatment Citation[18] and thus conclude that HT induces an apoptotic response. EGR1 is central for tumor growth, but has also been implicated in pro-apoptotic signaling Citation[27]. The effect of EGR1 is dependent on type of malignancy and stimulus Citation[27]. In human glioblastoma, constitutively expressed EGR1 suppresses growth and transformation Citation[28]. EGR1 induces growth arrest via expression of TGFß1, and signals apoptosis by p53 dependent and independent mechanisms Citation[27], Citation[29]. The pro-apoptotic effects of FOS, often co-regulated with EGR1, are dependent on wt p53 Citation[29] which was present in our tumor model. EGR1 interacts with the transcription factor JUN and augments its pro-apoptoticactivity Citation[27]. Phosphorylated EGR1 is also known to increase the expression of the pro-apoptotic protein ATF3 Citation[30]. It is intriguing that we found strong induction of all these genes in our study after treatment with HT. Several external stimuli are known to induce gene expression of ATF3, such as IR, UV and MMS Citation[31]. It is not surprising that heat also induces EGR1 and ATF3, but the precise mechanisms are not clear.
Transcription
The JUN, FOS, EGR1 and ATF3 genes discussed above are known transcription factors. They were members of two clusters found by GO mapping involved in regulation of transcription. We detected diverse responses in these groups, indicating that regulation of transcription by HT might be more specific than merely global up or down regulation of transcription.
Immune system
The biological process ‘immune response’ is a wide term (classified under ‘response to stimuli’), and our observations show a depression of the immune system in general. This depression was possibly directed toward the adaptive immune defense, especially lymphocytes, based on general depression of ‘lymphocyte activation’, ‘B cell receptor signaling pathway’, ‘T cell receptor signaling pathway’ and ‘T-cell homeostasis’, but also the innate immune response with general depression of mRNA levels of genes belonging to ‘Myeloid leukocyte activation’ and ‘Leukocyte chemotaxis’.
The literature is ambiguous concerning the effects of HT on the immune system, which are dependent on thermal dose (magnitude and duration of treatment), type of malignancy, model system (species), microenvironmental factors and the time of recovery before analysis Citation[32]. The results are more consistent in fever-range HT where enhancement of anti-tumor immune responses (mediated directly or via HSPs) and direct effects on the behavior of immune cells have been described Citation[13], Citation[16], Citation[17], Citation[33].
HSPs interact with APC cells that present antigens for cytotoxic T-lymphocytes Citation[16], Citation[17]. Three receptors for HSP on APC cells (Tlr2, CD14 and CD36) were detected at lower mRNA levels after HT, in contrast to the up regulated Lrp1. Hsp70 is a recognition structure for NK cells, and Hsp70 expression correlates with NK cell lysis Citation[15], Citation[34]. There is evidence that Klrd1 is a receptor on the NK cells responsible for Hsp70 targeting Citation[15]. Klrd1 heterodimers with other natural killer cell lectins like Klrc1 and Klrc2 Citation[15]. Klrd1, Klrb1b and CD69 are activating receptors on NK cells Citation[15], and it is interesting that they all exhibited reduced mRNA levels after HT. NK cells secrete cytokines like interferon γ (IFNγ), interleukin 2 (IL2), interleukin 12 (IL12), tumor necrosis factor α (TNFα) and Granulocyte-macrophage colony-stimulating factor (GM-CSF) without prior stimulation Citation[15]. We found a reduction in the mRNA levels of the IFNγ receptor and the IL2 receptor subunits after HT. IL2 affects growth and differentiation of a range of cells in the immune system as well as glioma cells. IL2Rγ is also an indispensable subunit of the functional IL21R complex which activates JAK1, JAK3, STAT1 and STAT3 Citation[35]. Furthermore, important members of the CD3 complex found in cytotoxic T-lymphocytes, chemokines and chemokine receptors were detected at reduced mRNA levels after HT.
These findings reveal a HT-induced suppression in mRNA levels of important receptors found in cells of the immune system, especially T-cells and NK cells. As these receptors have a central role in detection of Hsp70 (Tlr2, CD14, CD36, Klrd1), activation of NK cells (Klrd1, Klrb1b and CD69) and the subsequent signaling between immune cells (IFNγ receptor and IL2 receptor subunits), we hypothesize that there is a heat induced functional suppression of the immune system within tumors the first hours after treatment.
Immunohistochemical staining verified the presence of cytotoxic T-lymphocytes, NK-cells, B-lymphocytes and macrophages in the tumors. We detected a tendency towards increased numbers of these cells in tumors treated with HT, probably caused by some degree of inflammation in the periphery of the tumors. Consequently, the down regulated mRNA expression of immune cell associated genes was neither caused by a simple reduction in number of cells expressing these genes, nor by aberrant expression in tumor cells as the pure tumor cell culture only displayed minute mRNA levels of most of these genes. In a previous study, melanoma cells exposed to HT developed early resistance to cytotoxic T-cell lysis due to reduced antigen presentation by the tumor cells Citation[36]. A transient reduction of antigen presentation was found, but over time tumor cells remained susceptible to CTL recognition Citation[36]. In that study the immune cells were not exposed to HT. In our experiments, analyses of antigen presentation were outside the scope of the study. As described above, we reveal another mechanism in which the tumor cell immune system interplay may be suppressed, and further analyses will be done to disclose if this also is a transient phenomenon, or if the impact on the immune system is more long lasting. Only a few genes involved in the immune clusters found in vivo were also found in vitro as previously discussed. Therefore, further studies should be done in vivo in different malignancies, analyzing both gene expression and functional effects of the immune system and its interplay with tumor cells.
Blood vessels
Our study confirms that HT has impact on the angiogenesis of tumors. The underlying mechanisms for this effect of HT are not clear, and as diverse gene expression changes were detected in this cluster, we were not able to make further assumptions of the mechanisms involved.
Protein metabolism
We found clusters involved in protein modifications overrepresented after treatment with HT. Interference of heat on protein shape and stability is well known, and genes involved in protein folding were induced both in vivo and in vitro.
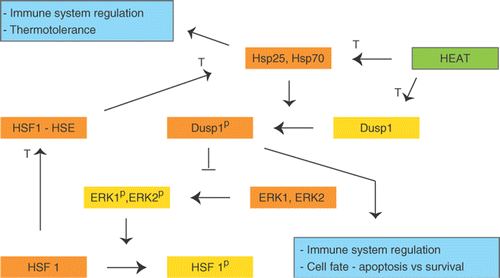
Amino acid phosphorylation is an important regulatory mechanism of proteins. Many signaling pathways (for instance MAPK signaling pathway) are sequentially activated by kinases. Analyses of our data revealed that genes involved in protein dephosphorylation were overrepresented after HT. Among these genes was a group DUSPs involved in dephosphorylation of central key proteins in signal transduction, such as the MAPK pathway. DUSP induction indicates inhibiting of central signal transduction pathways by HT. Three of the four DUSP genes detected by this GO cluster, DUSP19, DUSP8 and especially DUSP4, were strongly induced by HT in vivo. The fourth DUSP gene in this cluster, DUSP2, was strongly down regulated in vivo. Only one gene in this cluster, DUSP4, was also differentially expressed in vitro. A search among the 1,213 genes for additional DUSPs also revealed DUSP1. DUSP1 is important in a negative feedback loop of ERK1 (MAPK3) and ERK2 (MAPK1), and hence a key protein regulating their activity Citation[37], Citation[38]. ERK 1/2 were possibly activated in our tumors due to the mutation of BRAF. DUSP1 is also involved in regulation of immune function and cellular inflammatory responses Citation[37], Citation[39]. It is well documented that heat shock desensitizes the cells for new heat shock treatments (thermotolerance) lasting 2–3 days Citation[40]. However, the reports on influence of heat shock on other kinds of subsequent stresses are conflicting. Some report that heat shock still renders the cells responsive to a wide variety of other stress Citation[40], Citation[41], but most reports show that heat shock attenuates responses to other stresses as well, for instance inflammatory stimuli Citation[39], Citation[42], Citation[43]. In a monocyte cell line, attenuated response to an inflammatory stimulus subsequent to heat shock was conferred by induction of DUSP1, leading to loss of activation of ERK 1/2 and p38 by the inflammatory stimuli, and subsequent loss of inflammatory cytokines like TNFα Citation[43].
The tumors responded to heat shock by induced mRNA expression of both HSPs and DUSP1. These are potentially involved in a common regulatory loop (). DUSP1 is regulated in a delicate manner after HT and is proposed to play a crucial role in determining the cell fate Citation[44]. The activity of DUSP1 may initially be reduced by HT, but increases within an hour leading to increased rate of ERK 1/2 dephosphorylation Citation[39], Citation[44]. DUSP1 is phosphorylated by ERK 1/2, but may also be phosphorylated by Hsp27 and Hsp70 Citation[45], Citation[46]. DUSP1 dephosphorylates ERK 1/2, which then fails to phosphorylate Heat Shock Factor 1 (HSF-1). HSF-1 is active upon dephosphorylation and is transported to the nucleus where it induces the expression of Hsp27 and Hsp70, thereby creating a positive feedback loop Citation[46] (). Therapies inducing active DUSP1 may be an option for treatment of cancers, and one way to achieve this is heat induced high levels of HSPs.
Heat shock proteins
A panel of HSPs was upregulated after treatment with HT. The most prominent was Hsp70 (HSPA1B), reported to be one of the most termorespondent HSPs. HSPs are strong immune modulators, for reviews see Citation[13], Citation[15–17], Citation[32], Citation[33], Citation[47]. Enhancement of tumor antigen processing by increase of extra-cellular Hsp70 and increased susceptibility of NK-cells mediated tumor cell lysis have been reported Citation[13], Citation[15–17], Citation[34]. The HSPs also have effects inside the cells where they act as chaperons and inhibit apoptosis Citation[14], Citation[32], Citation[48]. Stimulation of the immune system by HSPs indicate HSPs as suitable agents for immunotherapy Citation[13], Citation[49] while their stimulation of growth and apoptosis implies that HSPs could be targets of therapy Citation[14].
DNAJB4 is a Hsp40 kD protein, recently postulated to be an inhibitor of invasion, metastasis and angiogenesis Citation[50], Citation[51]. In HeLa cells DNAJB4 had a peak in expression (2.9 times) six hours after HT treatment Citation[26]. We found similar results both in vivo and in vitro, which indicates that DNAJB4 expression in tumor cells is independent of the microenvironment.
Closing remarks
One gene may be capable of inducing several biological processes. It is hard to distinguish which of these processes have the most impact on tumor growth. Nevertheless, we are confident that the biological processes discussed in this study represent processes that are affected by HT, directly or indirectly. This study confirms previous knowledge of the importance of several biological processes in response to HT, but also sheds new light on these, for instance by indicating early down regulation of genes involved in the immune system by HT at 43°C. This may be important in treatment scheduling.
Acknowledgements
We thank Mette Pernille Myklebust and Solveig Angelskår for technical assistance.
The authors were supported by grants from The Norwegian Cancer Society: grant no. 88214 (O. Dahl), 89075/024 (E. D. Borkamo) and 06141/001 (Ø. Fluge), and from the L. Meltzers Foundation (E. D. Borkamo).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Dahl O, Overgaard J. Hyperthermia. Int:. Oxford textbook of oncology, J-C Hariot, R Souhami, I Tannock, P Hohenberger. Oxford University Press, Oxford 2002; 1: 511–525, Second edition
- Kampinga HH. Cell biological effects of hyperthermia alone or combined with radiation or drugs: A short introduction to newcomers in the field. Int J Hyperthermia 2006; 22: 191–196
- Kong G, Braun RD, Dewhirst MW. Hyperthermia enables tumor-specific nanoparticle delivery: Effect of particle size. Cancer Res 2000; 60: 4440–4445
- Ponce AM, Viglianti BL, Yu D, Yarmolenko PS, Michelich CR, Woo J, Bally MB, Dewhirst MW. Magnetic resonance imaging of temperature-sensitive liposome release: Drug dose painting and antitumor effects. J Natl Cancer Inst 2007; 99: 53–63
- Fajardo LF, Prionas SD, Kowalski J, Kwan HH. Hyperthermia inhibits angiogenesis. Radiat Res 1988; 114: 297–306
- Denekamp J. Vascular endothelium as the vulnerable element in tumours. Acta Radiol Oncol 1984; 23: 217–225
- Song CW. Effect of local hyperthermia on blood flow and microenvironment: A review. Cancer Res 1984; 44: 4721s–4730s
- Horsman MR. Tissue physiology and the response to heat. Int J Hyperthermia 2006; 22: 197–203
- Dewey WC. Failla memorial lecture. The search for critical cellular targets damaged by heat. Radiat Res 1989; 120: 191–204
- Lepock JR. How do cells respond to their thermal environment?. Int J Hyperthermia 2005; 21: 681–687
- Roti Roti JL, Kampinga HH, Malyapa RS, Wright WD, vanderWaal RP, Xu M. Nuclear matrix as a target for hyperthermic killing of cancer cells. Cell Stress Chaperones 1998; 3: 245–255
- Nakahata K, Miyakoda M, Suzuki K, Kodama S, Watanabe M. Heat shock induces centrosomal dysfunction, and causes non-apoptotic mitotic catastrophe in human tumour cells. Int J Hyperthermia 2002; 18: 332–343
- Calderwood SK, Theriault JR, Gong J. How is the immune response affected by hyperthermia and heat shock proteins?. Int J Hyperthermia 2005; 21: 713–716
- Coss RA. Inhibiting induction of heat shock proteins as a strategy to enhance cancer therapy. Int J Hyperthermia 2005; 21: 695–701
- Multhoff G. Activation of natural killer cells by heat shock protein 70. Int J Hyperthermia 2002; 18: 576–585
- Milani V, Noessner E, Ghose S, Kuppner M, Ahrens B, Scharner A, Gastpar R, Issels RD. Heat shock protein 70: Role in antigen presentation and immune stimulation. Int J Hyperthermia 2002; 18: 563–575
- Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol 2002; 2: 185–194
- Borkamo ED, Fluge Ø, Mella O, Akslen LA, Bruland O, Dahl O. Hyperthermia improves the antitumour effect of metronomic cyclophosphamide in a rat transplantable brain tumour. Radiother Oncol 2008 2008; 86(3)435–442, doi:10.1016/j.radonc.2008.01.022
- Makino H, Ishizaka Y, Tsujimoto A, Nakamura T, Onda M, Sugimura T, Nagao M. Rat p53 gene mutations in primary Zymbal gland tumors induced by 2-amino-3-methylimidazo[4,5-f]quinoline, a food mutagen. Proc Natl Acad Sci USA 1992; 89: 4850–4854
- Rollins LA, Leone-Kabler S, O'Sullivan MG, Miller MS. Role of tumor suppressor genes in transplacental lung carcinogenesis. Mol Carcinog 1998; 21: 177–184
- Swafford DS, Middleton SK, Palmisano WA, Nikula KJ, Tesfaigzi J, Baylin SB, Herman JG, Belinsky SA. Frequent aberrant methylation of p16INK4a in primary rat lung tumors. Mol Cell Biol 1997; 17: 1366–1374
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25: 25–29
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res 1996; 6: 986–994
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–5121
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. Mutations of the BRAF gene in human cancer. Nature 2002; 417(6892)949–954
- Kato N, Kobayashi T, Honda H. Screening of stress enhancer based on analysis of gene expression profiles: Enhancement of hyperthermia-induced tumor necrosis by an MMP-3 inhibitor. Cancer Sci 2003; 94: 644–649
- Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol 2002; 193: 287–292
- Huang RP, Liu C, Fan Y, Mercola D, Adamson ED. Egr-1 negatively regulates human tumor cell growth via the DNA-binding domain. Cancer Res 1995; 55: 5054–5062
- Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther 1998; 5: 3–28
- Yamaguchi K, Lee SH, Kim JS, Wimalasena J, Kitajima S, Baek SJ. Activating transcription factor 3 and early growth response 1 are the novel targets of LY294002 in a phosphatidylinositol 3-kinase-independent pathway. Cancer Res 2006; 66: 2376–2384
- Fan F, Jin S, Amundson SA, Tong T, Fan W, Zhao H, Zhu X, Mazzacurati L, Li X, Petrik KL, et al. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene 2002; 21: 7488–7496
- Milani V, Noessner E. Effects of thermal stress on tumor antigenicity and recognition by immune effector cells. Cancer Immunol Immunother 2006; 55: 312–319
- Ostberg JR, Repasky EA. Emerging evidence indicates that physiologically relevant thermal stress regulates dendritic cell function. Cancer Immunol Immunother 2006; 55: 292–298
- Multhoff G, Botzler C, Wiesnet M, Eissner G, Issels R. CD3- large granular lymphocytes recognize a heat-inducible immunogenic determinant associated with the 72-kD heat shock protein on human sarcoma cells. Blood 1995; 86: 1374–1382
- Asao H, Okuyama C, Kumaki S, Ishii N, Tsuchiya S, Foster D, Sugamura K. Cutting edge: The common gamma-chain is an indispensable subunit of the IL-21 receptor complex. J Immunol 2001; 167: 1–5
- Milani V, Frankenberger B, Heinz O, Brandl A, Ruhland S, Issels RD, Noessner E. Melanoma-associated antigen tyrosinase but not Melan-A/MART-1 expression and presentation dissociate during the heat shock response. Int Immunol 2005; 17: 257–268
- Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 2007; 26: 3203–3213
- Lin YW, Yang JL. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. J Biol Chem 2006; 281: 915–926
- Wong HR, Dunsmore KE, Page K, Shanley TP. Heat shock-mediated regulation of MKP-1. Am J Physiol Cell Physiol 2005; 289: C1152–C1158
- Dorion S, Landry J. Activation of the mitogen-activated protein kinase pathways by heat shock. Cell Stress Chaperones 2002; 7: 200–206
- Dorion S, Berube J, Huot J, Landry J. A short lived protein involved in the heat shock sensing mechanism responsible for stress-activated protein kinase 2 (SAPK2/p38) activation. J Biol Chem 1999; 274: 37591–37597
- Malhotra V, Wong HR. Interactions between the heat shock response and the nuclear factor-kappaB signaling pathway. Crit Care Med 2002; 30: S89–S95
- Sanlorenzo L, Zhao B, Spight D, Denenberg AG, Page K, Wong HR, Shanley TP. Heat shock inhibition of lipopolysaccharide-mediated tumor necrosis factor expression is associated with nuclear induction of MKP-1 and inhibition of mitogen-activated protein kinase activation. Crit Care Med 2004; 32: 2284–2292
- Yaglom J, O’Callaghan-Sunol C, Gabai V, Sherman MY. Inactivation of dual-specificity phosphatases is involved in the regulation of extracellular signal-regulated kinases by heat shock and hsp72. Mol Cell Biol 2003; 23: 3813–3824
- Brondello JM, Pouyssegur J, McKenzie FR. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science 1999; 286: 2514–2517
- Seo HR, Chung DY, Lee YJ, Lee DH, Kim JI, Bae S, Chung HY, Lee SJ, Jeoung D, Lee YS. Heat shock protein 25 or inducible heat shock protein 70 activates heat shock factor 1: Dephosphorylation on serine 307 through inhibition of ERK1/2 phosphorylation. J Biol Chem 2006; 281: 17220–17227
- Calderwood SK, Mambula SS, Gray PJ, Jr, Theriault JR. Extracellular heat shock proteins in cell signaling. FEBS Lett 2007; 581(19)3689–3694
- Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst 2000; 92: 1564–1572
- Wang XY, Li Y, Yang G, Subjeck JR. Current ideas about applications of heat shock proteins in vaccine design and immunotherapy. Int J Hyperthermia 2005; 21: 717–722
- Albini A, Pfeffer U. A new tumor suppressor gene: Invasion, metastasis, and angiogenesis as potential key targets. J Natl Cancer Inst 2006; 98: 800–801
- Tsai MF, Wang CC, Chang GC, Chen CY, Chen HY, Cheng CL, Yang YP, Wu CY, Shih FY, Liu CC, et al. A new tumor suppressor DNAJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst 2006; 98: 825–838