Abstract
Purpose: To analyze effects of hyperthermia on immune cells within tumors.
Materials and methods: Subcutaneously implanted rat gliomas were treated with water-bath hyperthermia for one hour at day zero (HT) (intratumoral temperature of 43°C), low-dose metronomic cyclophosphamide (CTX) 35 mg/kg three times a week for two weeks, both HT and CTX (CTX-HT0), or saline. Tumors harvested at day 1, 7, 14 and 21 were analyzed for changes in gene expression by microarrays, focusing on genes expressed in immune cells. Microarray analyses and real-time RT-PCR were previously performed on tumors harvested day zero at 0, 45, 90 and 180 minutes after end of treatment. Gene expression kinetics of selected genes were analyzed in all tumors by quantitative real-time RT-PCR (qPCR) to validate the results of the microarrays.
Results: Previous microarray analyses have indicated a suppression of gene expression in immune cells within tumors by hyperthermia the first three hours after treatment. qPCR analyses of CD14, CD36, Klrd1 (CD94), Tlr2 and Lrp1 confirmed these results. Global mRNA screen revealed no hyperthermia-induced changes in gene expression of immune cells at day 1, 7, 14 and 21. qPCR analyses of CD14, CD36, Klrd1 (CD94), Tlr2 and Lrp1 confirmed the results of the microarrays, i.e. none of these were differentially expressed after the first day, with the exception of Lrp1 which displayed elevated mRNA levels.
Conclusion: The suppression in gene expression of a range of immune cells within tumors treated with hyperthermia at 43°C is a transient phenomenon.
Introduction
We have recently analyzed global gene expression in tumors harvested the first three hours after treatment with local hyperthermia (mean intratumoral temperature of 43°C, one hour treatment), and revealed that hyperthermia suppressed gene expression in immune cells within tumors the first three hours after end of treatment Citation[1]. Several investigators have previously shown that fever-range hyperthermia (39–40°C) enhances anti-tumor immune responses. This is especially linked to induction of heat shock protein 70 (Hsp70), but direct effects on the behavior of the immune cells have also been reported Citation[2–5]. Hyperthermia has therefore been suggested as a potential adjuvant regimen to tumor vaccines based on Hsp70 Citation[2]. The contrasting results could indicate a differential effect on the immune system dependent on thermal dose (magnitude and duration of treatment), as also discussed by others Citation[6],Citation[7].
We used microarray analyses to elucidate whether or not suppression of gene expression in immune cells within tumors by hyperthermia at 43°C in our rat brain tumor model was a transient or long-lasting phenomenon. Rats were treated with hyperthermia with or without adjuvant low-dose cyclophosphamide (CTX) according to a regimen presented in detail elsewhere Citation[8]. Global mRNA analyses were performed on fresh frozen tissue samples harvested at day 1, 7, 14 and 21, and the kinetics of mRNA expression of some selected genes, CD 14 antigen (CD14), CD36 antigen (CD36), killer cell lectin-like receptor subfamily D member 1 (Klrd1) (alias CD94), toll-like receptor 2 (Tlr2) and low density lipoprotein receptor-related protein 1 (Lrp1) were analyzed by quantitative real-time RT-PCR (qPCR) to verify the results of the microarrays. Klrd1 is a receptor on natural killer (NK) cells responsible for detecting Hsp70. The other four genes encode receptors for Hsps on antigen presenting cells (APC). qPCR analyses were also performed on the tumor samples that had been sampled the first three hours after end of treatment at day zero and analyzed for changes in global gene expression by microarrays in a previous study Citation[1].
Materials and methods
Tumor tissue
Tissue was taken from tumors treated as previously described Citation[8]. Briefly, we used BDIX rats with a BT4An tumor (aggressive glioma) grown subcutaneously on the back leg. The experimental setup is shown in . Animals were allocated to the following groups: hyperthermia at 44.1° ± 0.1°C water bath (mean tumor temperature of 43°C) for one hour (HT); CTX 35 mg/kg intraperitoneal injection 3 times a week for a total of 6 treatments (CTX); combined therapy with hyperthermia immediately following the first injection of CTX (day zero) (CTX-HT0); or controls injected with saline. Two samples were collected from each group at each of the days 1, 7, 14 and 21. The rats were killed using CO2 gas, tumors harvested and frozen directly on nitrogen. Total RNA was extracted and stored at −80°C.
Table I. Flowchart for treatment and sample collection.
RNA purification and cDNA synthesis
Total RNA extraction and cDNA synthesis were performed according to standard protocols as previously described Citation[8]. Total RNA concentrations and quality were assessed using Nanodrop Spectrophotometer (Nanodrop Technologies, Wilmington, DE) and all samples (total RNA) had OD 260/280 in the range of 1.98–2.13. The samples were also assessed with 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Samples to be used for qPCR were DNase I treated (Ambion, Austin, TX). For cDNA reactions, inputs of 80 and 200 ng DNase I treated RNA respectively, were used for samples harvested the first day and subsequent days. Final volumes of cDNA reactions were 100 and 150 µl respectively. We used an input of 2 µg DIG-labeled cDNA product for microarray analyses.
Gene expression analyses
For analyses of global gene expression, Applied Biosystems Rat Genome Survey Microarray (Foster City, CA) analyzing 26.857 rat genes was used according to a previously described procedure Citation[1]. Files were imported as single channel data and analyzed with J-Express Pro 2.7 (Molmine AS, Bergen, Norway). qPCR was conducted as described elsewhere Citation[8], and the results analyzed using the standard curve method Citation[9]. All samples were expressed as ratios of relative amount of probe versus an internal control probe. As internal control, we used ß-actin (NM_031144) (Applied Biosystems Assay ID Rn00667869_m1). We investigated gene expression of CD14 (NM_021744) (Rn00572656_g1), CD36 (NM_031561) (Rn00580728_m1), Klrd1 (CD94) (NM_012745) (Rn01640401_m1), and Tlr2 (NM_198769) (Rn02133647_s1). All probes from Applied Biosystems.
Ethics
All the experiments and procedures described regarding the treatment of animals were approved by the local responsible laboratory animal science specialist under the surveillance of the Norwegian animal research authority. The experiments were conducted in accordance with the laws and regulations controlling experiments on live animals in Norway.
Statistics
Statistical analysis of microarrays (SAM) (calculated by J-express) was used to analyze the difference between tumors treated with CTX-HT0 or HT versus CTX or saline. The threshold for false discovery rate (FDR) was set to 2.5%. The FDR incorporates the uncertainty of false findings in large datasets. We reanalyzed the material with a paired SAM analysis of tumors treated with or without HT (HT versus C and CTX-HT0 versus CTX, tumors in each pair being harvested at the same time intervals) with the same results as the unpaired SAM analysis (data not shown). Individual genes were also analyzed with a Student's t test (J-express). For analyses of qPCR results, we sorted the tumors in two groups, one with samples from tumors treated with HT alone or the combination of HT and CTX (all time intervals), the other group with samples from tumors treated with CTX alone or saline (all time intervals). We analyzed the results with a Student's t test with the hypothesis that there is a difference between tumors treated with or without HT (α = 0.05). Real time RT-PCR statistics were performed using SPSS 14.02 statistical package (SPSS Inc., Chicago, IL).
Results
Tumors were sampled at day 1, 7, 14 and 21. Control tumors after day 7 and CTX alone and HT alone treated tumors after day 14 could not be sampled due to large tumor-burdens requiring the rats to be killed at these days. Two tumors were sampled from each group at each time interval. All tumors were gathered and sorted into two groups based on treatment with or without hyperthermia. SAM analysis revealed no change in gene expression based on hyperthermia treatment. We then selectively searched for a wide range of central genes in the immune system, previously found differentially expressed three hours after hyperthermia Citation[1] (). These genes are involved in the interplay between tumor cells, heat shock proteins (Hsps) and important cells in the immune system (lymphocytes, NK cells and APC cells). Individual statistical analysis using Student's t test confirmed that none of the genes were significantly differentially expressed with exception of chemokine receptor 5 (CCR5) which was 2.1 times up-regulated after hyperthermia (p = 0.036).
Figure 1. Supervised screen of microarrays for selected genes found in leucocytes. Selection based upon previous report of genes found differentially expressed first three hours after HT treatment Citation[1]. Two tumors harvested at each time interval, denoted a and b. Statistical analysis: statistical analysis of microarrays (SAM); HT/CTX-HT0 treated tumors compared to controls/CTX treated tumors; 26.857 genes analyzed; log2 expressed single channel data. Blank spots indicate missing data.
![Figure 1. Supervised screen of microarrays for selected genes found in leucocytes. Selection based upon previous report of genes found differentially expressed first three hours after HT treatment Citation[1]. Two tumors harvested at each time interval, denoted a and b. Statistical analysis: statistical analysis of microarrays (SAM); HT/CTX-HT0 treated tumors compared to controls/CTX treated tumors; 26.857 genes analyzed; log2 expressed single channel data. Blank spots indicate missing data.](/cms/asset/493e21d8-e182-4fda-861e-e24e7eed9ea9/ihyt_a_339962_f0001_b.gif)
To verify the microarray analyses from the previous study analyzing global gene expression the first three hours after hyperthermia Citation[1], we analyzed gene expression by qPCR of CD14, CD36, Klrd1, Tlr2 and Lrp1 (). When we compared HT/CTX-HT0 treated tumors with controls/CTX treated tumors, we detected reduced mRNA levels of CD14, CD36, Klrd1 and Tlr2 (p < 0.001) and increased levels of Lrp1 (p < 0.05). A more detailed analysis of the graphs displayed reduced gene expression of CD14 by both HT and CTX-HT0. Gene expression of CD36 was reduced by HT, increased by CTX, and stable after CTX-HT0. Klrd1 and Tlr2 mRNA levels were reduced by both HT, CTX and CTX-HT0 compared to controls. Gene expression of Lrp1 was increased by CTX-HT0.
Figure 2. Quantitative PCR results from tumors harvested 0, 45, 90 and 180 min after end of treatment the first day (day 0) and from tumors harvested at days 1, 7, 14 and 21. All samples are expressed as ratios of concentration of probe versus a control probe, adjusted so that the mean of the controls at the first time-point equals one. Statistical analysis: HT/CTX-HT0 treated tumors compared to controls/CTX treated tumors; **p < 0.01; *p < 0.05; Student's t test.
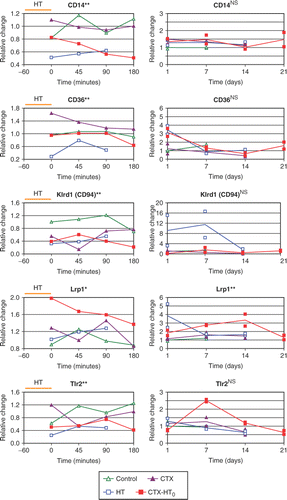
Quantitative PCR analyses of CD14, CD36, Klrd1, Tlr2 and Lrp1 were also performed on the tumors harvested 1–21 days following hyperthermia treatment. Lrp1 displayed a significantly increased mRNA level in HT/CTX-HT0 treated tumors compared to controls/CTX treated tumors. The other genes were not differentially expressed. If any change in gene expression, there was a tendency towards an increase in gene expression in subsequent days after treatment with HT or CTX-HT0 (increase in gene expression of CD36 1 day after HT and CTX-HT0, of Klrd1 1 and 7 days after HT, and of Tlr2 7 days after CTX-HT0) ().
Discussion
The SAM analysis revealed no genes differentially expressed on day 1–21 after treatment with hyperthermia. This analysis was conducted after all tumor samples had been gathered in two groups discriminating tumors treated with or without hyperthermia. This analysis detects changes in gene expression that are stably affected by hyperthermia the first three weeks after treatment, and is the same method that was used in a previous study where we reported a suppression in gene expression of immune cells within tumors the first three hours after treatment Citation[1]. Due to this choice of method, we set the FDR threshold in the SAM analysis in both studies at a strict value of 2.5%. This means that there is a 2.5% chance that the results are random findings. SAM analysis revealed no differential gene expression due to hyperthermia treatment between day 1 and 21. This is supported by the selective screen of central genes found in leucocytes where only CCR5 was found differentially expressed by Student's t test which is a less restrictive test than the SAM analysis.
Quantitative PCR analysis of CD14, CD36, Klrd1, Tlr2 and Lrp1 were in agreement with the previously published microarray results Citation[1] which indicated an early suppression in gene expression of immune cells within hyperthermia-treated tumors. The non-differential effect by CTX-HT0 treatment on CD36 mRNA seems to be caused by the fact that CTX induce CD36 while HT alone suppresses CD36. Quantitative PCR analysis of CD14, CD36, Klrd1, Tlr2 and Lrp1 of tumors harvested on subsequent days did not detect reduced mRNA levels after HT/CTX-HT0, but in fact indicated up-regulation of CD36, Klrd1, Tlr2 and Lrp1 mRNA levels at some time intervals after HT and CTX-HT0.
An early resistance to cytotoxic T-cell (CTL) lysis due to reduced antigen presentation has previously been reported in melanoma cells exposed to hyperthermia. This was a transient phenomenon that only lasted hours after treatment Citation[10]. The activity of NK cells is regulated in a delicate balance between activating and inhibitory receptors and it seems clear that hyperthermia, dependent on temperature and duration, affects the number and or the activity of these cells Citation[6],Citation[7]. Hyperthermia-induced changes in number of immune cells and in their activity have primarily been investigated in rodents and humans treated with whole body hyperthermia or in cell cultures. Consistent results have mainly emerged after whole body hyperthermia where enhancement in NK cell numbers and activity has been reported Citation[7]. Some mechanisms were suggested, but much remains to be elucidated. Our previous results indicated a reduction in gene expression of immune cells within tumors, including NK cells, by 1-h local water-bath hyperthermia Citation[1]. However, we now disclose that this phenomenon seems to be transient. Furthermore, our results indicate that if any impact on leucocytes within tumors later than one day after hyperthermia, it is more likely to be stimulatory. Others have observed corresponding effects using in vivo ultrasound-generated hyperthermia on hamster spleen, detecting depression of NK cell cytotoxicity and lymphopenia at 4 hours, recovering within respectively 24 and 48 hours Citation[11]. Whole body hyperthermia and local hyperthermia tend to have differential impact on immune cells and on the interplay between tumor cells and immune cells. Healthy human subjects treated with whole body hyperthermia, but not local hyperthermia, increased NK cell activity Citation[12]. We show a rapid but transient suppression of gene expression in immune cells within tumors by local hyperthermia. Our findings are further supported by a recent report from Funaki et al. Citation[13]. They reported a transient reduced cytotoxicity of immune cells exposed to hyperthermia at 43°C. Consistent evidence emerges for a short-lived suppression of immune cells by local hyperthermia, but the functional relevance of this phenomenon should be further assessed in different malignancies. Comparative in vivo and in vivo models systems would be favorable to disclose the impact of intact tumor/immune cell microenvironment and cytokine signaling. A more complete characterization of infiltrating cells in the tumor bed is needed Citation[7], and gene- and protein expression of immune cells within tumors should be compared with those of the blood. In doing so, it is important to keep in mind that treating a patient with local hyperthermia probably yields a fever-range temperature response in the rest of the body (core body temperature increase during therapy). This might actually be favorable with the probability to exploit both the effects of local hyperthermia on tumor blood flow, angiogenesis and tumor cell cytotoxicity, as well as the immune stimulating effects of whole body fever range hyperthermia. Our results suggest that the suppression of immune cells by local hyperthermia is transient and therefore does not hinder the combination of hyperthermia at 43°C and immunotherapy with for instance Hsp vaccines Citation[14] the following day. Both the early and the long-term biological responses to hyperthermia are important to elucidate in detail since these may affect the way we combine hyperthermia with other treatment modalities. A combination of local hyperthermia and metronomic chemotherapy inducing antiangiogenic and apoptotic responses combined with the effect of whole body fever range hyperthermia and Hsp vaccines might be a powerful and complementing regimen.
Acknowledgements
We thank M. P. Myklebust and S. Angelskår for technical assistance. Supported by grants 88214 (O. Dahl), 89075/024 (E. D. Borkamo) and 06141/001 (Ø. Fluge) from The Norwegian Cancer Society and L. Meltzers Foundation (E.D. Borkamo).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Borkamo ED, Dahl O, Bruland O, Fluge Ø. Global gene expression analyses reveal changes in biological processes after hyperthermia in a rat glioma model. Int J Hyperthermia 2008; 24(5)425–441
- Calderwood SK, Theriault JR, Gong J. How is the immune response affected by hyperthermia and heat shock proteins?. Int J Hyperthermia 2005; 21: 713–716
- Milani V, Noessner E, Ghose S, Kuppner M, Ahrens B, Scharner A, Gastpar R, Issels RD. Heat shock protein 70: Role in antigen presentation and immune stimulation. Int J Hyperthermia 2002; 18: 563–575
- Srivastava P. Roles of heat-shock proteins in innate and adaptive immunity. Nat Rev Immunol 2002; 2: 185–194
- Ostberg JR, Repasky EA. Emerging evidence indicates that physiologically relevant thermal stress regulates dendritic cell function. Cancer Immunol Immunother 2006; 55: 292–298
- Milani V, Noessner E. Effects of thermal stress on tumor antigenicity and recognition by immune effector cells. Cancer Immunol Immunother 2006; 55: 312–319
- Dayanc BE, Beachy SH, Ostberg JR, Repasky EA. Dissecting the role of hyperthermia in natural killer cell mediated anti-tumor responses. Int J Hyperthermia 2008; 24: 41–56
- Borkamo E, Fluge O, Mella O, Akslen LA, Bruland O, Dahl O. Hyperthermia improves the antitumour effect of metronomic cyclophosphamide in a rat transplantable brain tumour. Radiother Oncol 2008; 86: 435–442
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res 1996; 6: 986–994
- Milani V, Frankenberger B, Heinz O, Brandl A, Ruhland S, Issels RD, Noessner E. Melanoma-associated antigen tyrosinase but not Melan-A/MART-1 expression and presentation dissociate during the heat shock response. Int Immunol 2005; 17: 257–268
- Johnston RL, Rao GR, Tompkins WA, Cain CA. Effects of in vivo ultrasound hyperthermia on natural killer cell cytotoxicity in the hamster. Bioelectromagnetics 1986; 7: 283–293
- Blazickova S, Rovensky J, Koska J, Vigas M. Effect of hyperthermic water bath on parameters of cellular immunity. Int J Clin Pharmacol Res 2000; 20: 41–46
- Funaki J, Kokura S, Okayama T, Adachi S, Nakune K, Takeda A, Shimabukaro T, Takeda T, Kondo M, Yoshikawa T. The efficacy of hyperthermia combined with cancer immunotherapy. 10th International Congress on Hyperthermic Oncology, Klinikum Grosshadern, University of MunichGermany, 2008
- Wang XY, Li Y, Yang G, Subjeck JR. Current ideas about applications of heat shock proteins in vaccine design and immunotherapy. International Journal of Hyperthermia 2005; 21: 717–722
