Abstract
Hypoxia inducible factor (HIF), a master regulator of critical genes for cell survival under hypoxic conditions, is known to be related to tumorigenesis and progression of renal cell carcinoma. N-methylpyrrole (Py)-N-methylimidazole (Im) hairpin polyamides are synthetic organic compounds that recognize and bind to the minor grooves of specific DNA sequences. We synthesized three Py-Im hairpin polyamides targeting the flanking sequences of hypoxia responsive element (HRE; a binding site of HIF) in the promoter region of the vascular endothelial growth factor (VEGF) gene. The effects of the polyamides on HIF-induced transcription were evaluated by a luciferase assay using a reporter plasmid containing a VEGF promoter. Real time reverse-transcriptase polymerase chain reaction and enzyme-linked immunosorbent assay were performed to examine the effects of the polyamides on the transcription and secretion of VEGF in A498 renal cell carcinoma cells, which have a frame-shift mutation in the von Hippel-Lindau gene. A combination of three Py-Im hairpin polyamides suppressed HIF-induced transcription in reporter assays using 293 cells and successfully suppressed transcription and translation of the VEGF gene in A498 cells. Inhibition of the HIF-HRE interaction was confirmed by an electrophoresis mobility shift assay. An approach using Py-Im hairpin polyamides may be a new strategy for the treatment of renal cell carcinoma.
Hypoxia inducible factor (HIF) is a key regulator of molecules that are critical to cell survival under hypoxic conditions Citation[1]. In low-oxygen conditions, HIFs (HIF-1 or HIF-2 bind to the hypoxia responsive element (HRE), and trigger the transcription of target genes such as vascular endothelial growth factor (VEGF) or erythropoietin. Under normal oxygen tension, the von Hippel-Lindau (VHL) protein binds to the alpha subunits of the HIFs (HIF-1α or HIF-2α) and facilitates their rapid degradation.
Renal cell carcinoma is characterized by extensive neovascularization. Recently, frequent mutations of the VHL gene have been demonstrated in familial and sporadic renal cell carcinoma with clear cell phenotypes. Stabilization of HIFs due to an impaired VHL-HIF interaction has been shown to be an underlying mechanism of hypervascularity in renal cell carcinoma Citation[2]. Thus, suppression of transcriptional activity of these HIFs may lead to decreases in de novo vascular formation and provide a new treatment modality for renal cell carcinoma that is resistant to radiation or chemotherapy.
N-methylpyrrole (Py)-N-methylimidazole (Im) hairpin polyamides have been shown to bind to the minor groove of specific DNA sequences with affinity and specificity similar to those of transcriptional factors Citation[3]. Successful suppression of transcription by Py-Im polyamides has been demonstrated for several genes Citation[4]. In the current study, we synthesized three Py-Im hairpin polyamides against sequences flanking the HRE of the human VEGF gene. A combination of three Py-Im hairpin polyamides suppressed HIF-induced transcription in reporter assays and successfully suppressed transcription and translation of the VEGF gene in A498 cells.
Materials and methods
Determination of target sequences
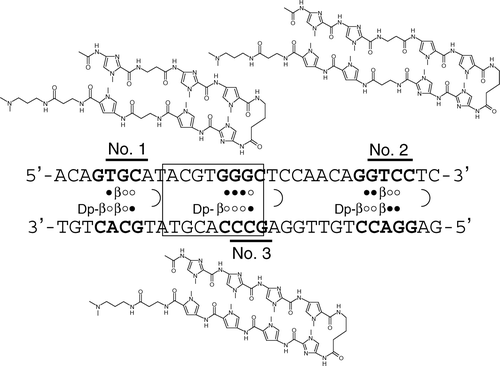
On the basis of the rules of sequence-recognition by Py-Im polyamides Citation[4–6], we designed three Py-Im hairpin polyamides targeting 4–5-base-pair sequences flanking the HRE of the human VEGF gene (). Target sequences were set between A/T or T/A pairs according to the reported sequences that successfully suppressed other transcriptional factors Citation[4]. For A/T or T/A, we used a β-alanine pair that degenerately recognizes these base pairs and increases the affinity of the polyamides to the target sequences by correcting intramolecular distortion Citation[7].
Figure 1. Target sequences and chemical structures of the N-methylpyrrole-N-methylimidazole hairpin polyamides. Consensus binding site for HIF is indicated by an open box. Open and closed circles represent imidazole and pyrrole rings, respectively. Dp and b denote dimethylaminopropylamine and b-alanine residues, respectively.
Preparation of pyrrole-imidazole hairpin polyamides
N-methylpyrrole (Py)-N-methylimidazole (Im) polyamides were synthesized by using a previously described Fmoc solid-phase method Citation[8]. Polyamides were purified by high performance liquid chromatography (HPLC) with a Jasco PU-980 HPLC pump, a UV-975 HPLC UV/VIS detector, and a Chemcobond 5-ODS-H column (4.6 mm×150 mm). HPLC was performed using 0.1% acetic acid and a linear gradient (20–30% for polyamide no. 1, 10–50% for polyamide no. 2, and 20–30% for polyamide no. 3) of acetonitrile at a flow rate of 1.0 ml/min with detection at 254 nm. Polyamides were eluted at 30.0 min. Structures of the synthesized polyamides were verified by electron spray ionization mass spectra (ESIMS) recorded on a PE Sciex API 165 mass spectrometer. Molecular weights of polyamides measured by ESIMS are listed in . Synthesized polyamides were lyophilized and stored at −20°C, and dissolved in dimethylsulfoxide for the experiments.
Table I. Molecular weights of polyamides measured by ESIMS.
Cells and culture conditions
A498 is an established cell line of undifferentiated renal cell carcinoma (American Type Culture Collection, Manassas, VA, USA) that lacks normal expression of the von Hippel-Lindau gene because of a frame-shift mutation at codon 213 Citation[9]. High-level expressions of HIF-1α and HIF-2α in A498 have been demonstrated Citation[10]. 293 is an immortalized human fetal kidney epithelial cell line with very low levels of HIF-1α and HIF-2α under normoxic conditions Citation[10]. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma Aldrich, St. Louis, MO, USA) containing 10% fetal bovine serum in a humid chamber with 5% CO2 at 37°C.
Plasmids
The following plasmids were used to evaluate the effects of the hairpin polyamides on HIF-1α-induced transcription: p(HA)HIF-1α (401Δ603), HA-tagged full-length human HIF-1α with internal deletion of oxygen-dependent degradation domain Citation[11–13]; pTRE-EPAS, full-length human HIF-2α Citation[10]; pARNT, full-length human ARNT Citation[11–13]; pGL3VEGF, pGL3 promoter (Promega, Madison, WI, USA) containing the 385-bp VEGF promoter sequence (−1175 to −790) Citation[14]; pCMVβ (BD biosciences, Palo Alto, CA, USA), a full-length human β-galactosidase cDNA.
Reporter assay
Two hundred and ninety three cells were seeded in 12-well plates (15×104 cells per well) and cultured in a humid chamber (5% CO2). Twenty-four hours later, 0.25 µg of pGL3VEGF, 0.1 µg of p(HA)HIF1α(401Δ 603) or pTRE-EPAS, and 0.1 µg of pARNT were transiently transfected using the FuGene transfection reagent (Roche Applied Science, Penzberg, Germany) according to the manufacturer's protocol. pCMVβ (0.1 µg) was also transfected as a reference plasmid. Two hours later, synthesized polyamides were added to the culture. Doses and combinations of the polyamides are shown in the figures. After incubation for another 24 h, cells were lyzed with M-PER reagent (Pierce, Rockford, IL, USA). Luciferase activity was determined with the Bright-Glo™ Luciferase assay system (Promega, Madison, WI, USA). The luminescence signal was evaluated by using a Gene Light 55 luminometer (Microtec, Tokyo, Japan). The activity of β-galactosidase was measured by using a mammalian β-galactosidase assay kit (Pierce, Rockford, IL, USA) and an MT Max microplate reader (Wako, Tokyo, Japan). Assays were carried out in triplicate and repeated at least twice. The results were expressed as a ratio of luciferase activity to β-galactosidase activity.
Real-time reverse transcriptase-polymerase chain reaction
A498 cells were seeded in 6-well plates (30×104/well). The medium was replaced 24 h later and a polyamide mixture (5 µM in total) was added to the culture. Total RNA was extracted from the cells after 24, 48, and 72 h using ISOGEN (Nippon Gene, Tokyo, Japan), an RNA-isolating reagent. cDNA was synthesized from 5 µg of the total RNA samples using the SuperScript™ First-Strand Synthesis System for reverse transcriptase-polymerase chain reaction (RT-PCR; Invitrogen, Carlsbad, CA, USA). Five microliters of 500-fold-diluted cDNA samples were subjected to real time PCR by using a Light Cycler™ real time PCR instrument (Roche Applied Science, Penzberg, Germany). Light Cycler™ primer sets for human VEGF or human GAPDH (Serach LC, Heidelberg, Germany) were used together with LightCycler FastStart DNA Master SYBR Green I (Roche Applied Science, Penzberg, Germany). Results were expressed as a ratio of VEGF mRNA to GAPDH mRNA. Experiments were carried out in triplicate and repeated at least twice.
Enzyme-linked immunosorbent assay
A498 cells were plated on 24-well plates (5×104/well). The medium was replaced 24 h later and a polyamide mixture (5 µM in total) was added to the culture. Conditioned media were collected after 24, 48 and 72 h. The concentration of VEGF in the conditioned media was measured by using a Cytokine Enzyme-Linked Immunosorbent Assay (ELISA) Human VEGF Kit (American Research Products, Belmont, MA, USA). Assays were carried out in triplicate and repeated at least twice. Results were expressed as a ratio of VEGF concentration to total protein concentration, which was determined by using the BCA™ Protein Assay Kit (Pierce, Rockford, IL, USA).
Cell viability assay
A498 or 293 cells were seeded in 24-well plates. The medium was replaced 24 h later and polyamides at designated concentrations were added to the culture. After 24 h of incubation, the viability of the cells was assessed by using a CellTiter 96 AQueous One Solution Cell Proliferation Assay reagent (Promega, Madison, WI, USA). For A498, viability at 48 and 72 h was also measured. All assays were carried out in triplicate and repeated at least twice.
Electrophoresis mobility shift assay
Nuclear extract (100 µl) was prepared from 1×106 A498 cells using the NE-PER Nuclear and Cytoplasmic Extraction Reagent (Pierce, Rockford, IL, USA) according to the manufacturer's instructions. The binding reaction was carried out by using a Light-Shift Chemiluminescent Electrophoresis Mobility Shift Assay (EMSA) Kit (Pierce, Rockford, IL, USA). Twenty femtomoles of a biotin-labeled double-stranded oligonucleotide (5′-GTGCATACGTGGGCTCCAACAGGTCC-3′) corresponding to the sequence harboring the HRE of the human VEGF gene was mixed with 10 nmol of the polyamide mixture (no. 1, 2 or 3) or a polyamide targeting the human telomere sequence (TTAGGG) Citation[15] in 20 µl of 1x binding buffer containing 2.5% glycerol, 5 mM MgCl2, 50 ng/µl poly(dIdC), and 0.05% NP40. The reaction mixture was incubated for 20 min at room temperature, mixed with 2 µl of the A498 nuclear extract, and then incubated for another 20 min at room temperature. The nuclear extract was substituted with an equal volume of H2O in a negative control. In a positive control, polyamide samples were replaced by an equal volume of H2O. The reaction products were mixed with 5x loading buffer, electrophoresed in a 6% polyacrylamide gel (Invitrogen, Carlsbad, CA, USA) in 0.5x Tris-borate-EDTA (TBE), and transferred to a nylon membrane (Pierce, Rockford, IL, USA). Biotin-labeled DNA was detected by using a Chemiluminescent Nucleic Acid Detection Module (Pierce, Rockford, IL, USA) according to the manufacturer's instructions and analyzed by using an LAS-1000 imaging system (Fujifilm, Tokyo, Japan).
Results
Dose-dependent suppression of HIF-1α-induced transcription by polyamide mixture
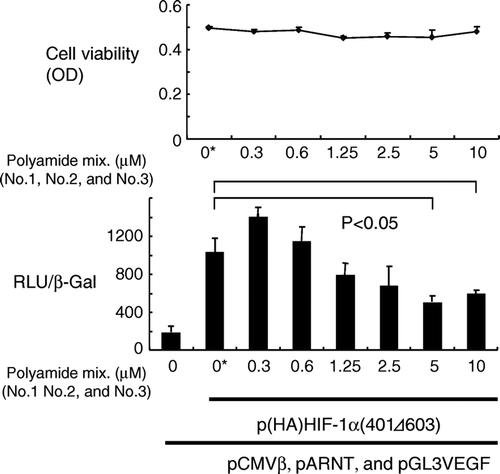
We evaluated the effects of the synthesized polyamides on HIF-1α -induced transcription by using a reporter assay. A luciferase-expressing vector with the promoter region of the human VEGF gene was used as a reporter plasmid. Because HIF-1α is fairly unstable under normal oxygen tension, we used a stable HIF-1α construct that is transcriptionally active. Because HIF-1α makes a heterodimer with ARNT (also known as HIF-1α) for binding to the HRE, a plasmid clone of the human ARNT gene was transfected along with the HIF-1α. A plasmid containing the human β-galactosidase gene was also transfected and used as a reference standard. As shown in , a combination of three hairpin polyamides suppressed transcription induced by HIF-1α in a dose-dependent manner to 56% of the control (vehicle only). The viability of the cells was not affected.
Figure 2. Dose-dependent inhibition of HIF-1α-induced transcription in 293 cells. Cells were transfected with plasmids coding hypoxia-stable HIF-1α, ARNT, and β-galactosidase, along with a reporter plasmid (pGL3VEGF), and then treated with the mixture of three pyrrole-imidazole hairpin polyamides for 24 h. Transcriptional activity was measured and expressed as a ratio of luciferase to β-galactosidase activity. Statistically significant suppression of transcription was noted at a concentration of 5 µM or greater. Cell viability was not affected by the polyamides. RLU and β-Gal denote relative luciferase units and β-galactosidase activity, respectively. OD: optical density. *: Vehicle (dimethylsulfoxide) only.
Suppression of HIF-1α-induced transcription by a combination of the three hairpin polyamides
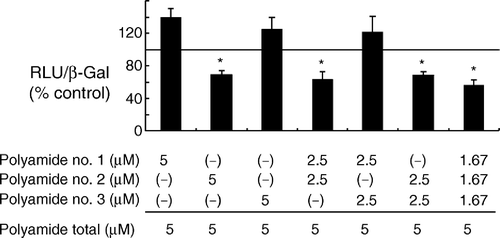
The effects of each polyamide alone and various combinations of the three polyamides were evaluated by using a reporter assay. As shown in , of the three polyamides, no.2 was the most potent suppressor of HIF-induced transcription at a concentration of 5 µM. A combination of all three polyamides, however, resulted in satisfactory suppression at lower concentrations of each polyamide compared with the monotreatment.
Figure 3. Effects of combinations of the three polyamides on HIF-1α -induced transcription in 293 cells. Cells were transfected with plasmids coding for hypoxia-stable HIF-1α(p(HA)HIF-1α(401Δ603)), ARNT (pARNT), and β-galactosidase (pCMVβ), along with a reporter plasmid (pGL3VEGF), and then incubated with a particular combination of the pyrrole-imidazole polyamides for 24 h. Transcriptional activity was expressed as a ratio of luciferase to β-galactosidase activity. A statistically significant reduction in transcription was achieved by applying all three Py-Im hairpin polyamides. A similar suppression was observed when polyamide no. 3 was applied alone or together with either polyamide no. 1 or no. 2. Cell viability was not affected by the treatment. RLU and β-Gal represent relative luciferase units and β-galactosidase activity, respectively. *: P < 0.05 compared with controls.
Suppression of HIF-2α-induced transcription by the polyamide mixture
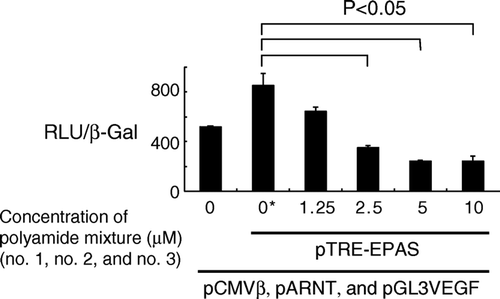
We also examined the effects of the polyamide mixture on HIF-2α-induced transcription by using a reporter assay. A luciferase-expressing vector with the promoter region of the human VEGF gene was used as a reporter plasmid. As in the experiments using HIF-1α, a plasmid clone of the human ARNT gene was co-transfected with a full-length clone of the human HIF-2α gene. A plasmid containing the human β-galactosidase gene was also transfected and used as a reference standard. As shown in , a combination of the three hairpin polyamides suppressed transcription induced by HIF-2α in a dose-dependent manner to 28% of the control (vehicle only).
Figure 4. Dose-dependent inhibition of HIF-2α-induced transcription in 293 cells. Cells were transfected with plasmids coding full-length HIF-2α (pTRE-EPAS), ARNT(pARNT), and β-galactosidase (pCMVβ), along with a reporter plasmid (pGL3VEGF), and then treated with a mixture of the three pyrrole-imidazole hairpin polyamides for 24 h. Transcriptional activity was measured and expressed as a ratio of luciferase to β-galactosidase activity. Statistically significant suppression of transcription was noted at a concentration of 2.5 µM or greater. RLU and β-Gal denote relative luciferase units and β-galactosidase activity, respectively. *: Vehicle (dimethylsulfoxide) only.
Suppression of transcription and secretion of VEGF in renal cell carcinoma cells by the polyamide mixture
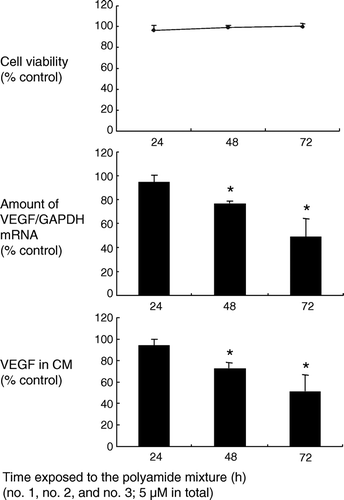
We examined the effects of the mixture of all three polyamides on VEGF transcription in renal cell carcinoma cells by using real-time RT-PCR. GAPDH mRNA was used as an internal standard. We used A498, a cell line derived from human renal cell carcinoma, with a frame-shift mutation in the VEGF gene. As shown in , the polyamide mixture at a concentration of 5 µM inhibited VEGF transcription in A498 cells in a time-dependent manner. An approximately 50% reduction in VEGF transcription relative to the control (vehicle only) was observed when cells were exposed to the polyamide mixture for 72 h. Inhibition of VEGF at the protein level was examined by ELISA using conditioned media. The results were consistent with those of real-time RT-PCR. Secretion of VEGF was suppressed by the polyamide mixture in a time-dependent manner. The amount of VEGF in the conditioned media was reduced to approximately 50% of the control (vehicle only) by exposing A498 cells to the polyamide mixture for 72 h. The viability of A498 cells was not affected by incubation with the Py-Im polyamides.
Figure 5. Suppressive effects of the mixture of three Py-Im hairpin polyamides on VEGF transcription and secretion in A498, a renal cell carcinoma cell line. Cells were treated with the mixture of three pyrrole-imidazole polyamides (5 mM in total) for 24–72 h. Transcription of the VEGF gene was evaluated by real time RT-PCR. The results were expressed as a ratio of VEGF mRNA to GAPDH mRNA. An approximately 50% reduction in transcription was observed after 72 h treatment. Consistently, VEGF secreted in conditioned medium (CM) decreased by approximately 50% compared with the control. *: P < 0.05 compared with controls.
Specific inhibition of HIF-HRE binding by the polyamide mixture
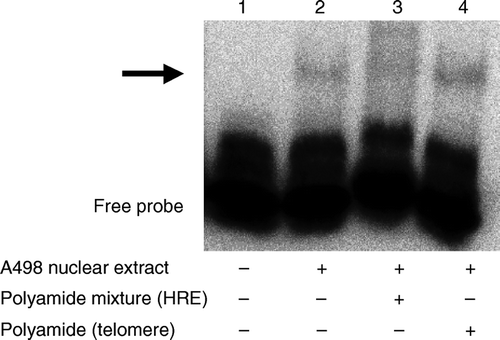
EMSA was carried out to confirm the inhibitory effects of the polyamide mixture on HIF-HRE binding (). Addition of the A498 nuclear extract to the reaction mixture caused an apparent mobility shift of the biotin-labeled DNA, which was suppressed by the polyamide mixture recognizing the human HRE. The mobility shift was not affected by a polyamide recognizing the human telomere sequence, which was used as a control.
Figure 6. Inhibitory effects of pyrrole-imidazole hairpin polyamides on the HIF-HRE interaction were evaluated by using an electrophoresis mobility shift assay. Nuclear extract of A498 human renal cell cancer cells caused a mobility shift (arrow) of a biotin-labeled double-stranded oligonucleotide corresponding to human HRE (lanes 1 and 2). The polyamide mixture against the HRE suppressed the mobility shift (lane 3), which was not affected by a polyamide recognizing the human telomere sequence (lane 4).
Statistics
Each assay was carried out in triplicate and repeated at least twice. Statistical significance was analyzed by using Student's t-test.
Discussion
VEGF transcription in renal cell carcinoma cells was effectively suppressed by the Py-Im hairpin polyamides in the current study. Inhibition of the HIF-HRE interaction was confirmed by EMSA to be the underlying mechanism of this suppression. The hairpin polyamide strategy was pioneered by Dervan and colleagues Citation[3]. Py and Im covalently link side-by-side in an anti-parallel arrangement that has been shown to recognize and bind to specific DNA sequences Citation[4–6]. Im-Py recognizes G:C, whereas Py-Im recognizes C:G. The Py-Py pair recognizes A:T or T:A base pairs. The hydroxypyrrole and Py pair (Hp–Py) distinguishes T:A from A:T and vice versa. Placing a β-alanine pair, which degenerately binds to T:A or A:T, increases the affinity and specificity for sequences by relaxing the rigid curvature of polyamides Citation[7]. Following these rules, regulation of gene expression by Py-Im hairpin polyamides has been successfully demonstrated for a variety of transcriptional factors, including Ets-1, TATA box (TBP) binding protein and lymphoid enhancer factor (LEF) 1 Citation[4]. Our data suggest that HIF-induced transcription can also be controlled effectively by competitive binding of multiple Py-Im hairpin polyamides.
In the current study, we prepared three polyamides recognizing 4–5 base pairs flanking the HRE of the VEGF gene, and administered them together to obtain maximum specificity. Because of technical constraints, it is difficult to synthesize long polyamide chains. Moreover, the longer the target sequence is, the greater the intramolecular distortion becomes. Thus, Py-Im hairpin polyamides against 4–5-base-pair sequences have been used in most studies. Recently, a hairpin polyamide that recognizes TACG within the HRE has been reported to inhibit VEGF transcription Citation[16]. Treatment with a combination of multiple polyamides is another option, and may increase the efficacy of treatment with Py-Im hairpin polyamides.
Py-Im hairpin polyamides have advantages as modulators of gene expression Citation[4]. First, their low molecular weight allows them to permeate through cell membranes: they simply pass through cell membranes and reach their target sites without special aids or vehicles such as expression vectors or liposomes. Designing hairpin polyamides is not difficult, and automated solid-phase synthesis is possible. Moreover, they have flexible sites for covalent attachment to other molecules. Hairpin polyamides linked with an alkylating agent may broaden the targets of transcriptional regulation of coding sequences Citation[17]. In fact, we have successfully inhibited gene transcription of the green fluorescent protein and luciferase by alkylating hairpin polyamides Citation[18].
Inhibiting the HIF transcription pathway is a fascinating strategy for cancer control, especially in the case of renal cell carcinomas. HIF controls genes related to the progression of cancers. Genes for platelet-derived growth factor (PDGF), VEGF, epidermal growth factor (EGF), and transforming growth factor-1α (TGF-1α are among those regulated by HIF-1 (a dimer of HIF-1α and ARNT). Therapeutic approaches using each of these molecules are being trialled clinically Citation[19]. Avastin™ (anti-VEGF antibody), SU11248 (a small molecule targeting the tyrosine kinase domain of the VEGF receptor), and Tarceva™ or Iressa™ (molecules targeting the tyrosine kinase domain of the EGF receptor) are currently being investigated as potential agents against advanced renal cell carcinoma. Recently, we have shown that glucocorticoids can down-regulate VEGF in renal cell carcinoma cells Citation[20]. Suppressing the binding of HIF-1 to the HRE may be a useful strategy for inhibition of these molecules. In addition, we and several other groups have demonstrated that HIF-2α, another α subunit of HIF, is more critical than HIF-1α in renal cell cancer Citation[10], Citation[21]. As HIF-2 (a dimer of HIF-2α and ARNT) shares an HRE with HIF-1, targeting the HIF-HRE interaction may be more useful in the management of renal cell carcinoma than using the molecular therapeutic drugs described earlier. In the current study, the polyamide mixture successfully suppressed transcription induced not only by HIF-1α but also by HIF-2α. Our findings may facilitate the realization of the “transcription therapy” concept proposed by Dervan Citation[5] for cancer treatment.
Conclusions
VEGF transcription was successfully suppressed by a combination of three Py-Im hairpin polyamides targeting the HRE. The use of Py-Im hairpin polyamides may be a new strategy for the treatment of renal cell carcinoma.
We thank Dr. Minori Koshiji and Ms. Kaoru Ito for their technical support. This work was supported by a Grant-in-Aid (14370505, 17591666, 17591667) for Scientific Research from the Ministry of Education, Science and Culture, Japan.
References
- Huang LE, Bunn HF. Hypoxia inducible factor and its biomedical relevance. J Biol Chem 2003; 278: 19575–8
- Kaelin WG. The von Hippel-Lindau tumor suppressor gene and kidney cancer. Clin Cancer Res 2004; 10: 6290–5s
- White S, Szewczyk JW, Turner JM, Baird EE, Dervan PB. Recognition of the four Watson-Crick base pairs in the DNA minor groove by synthetic ligands. Nature 1998; 391: 468–71
- Murty MSRC, Sugiyama H. Biology of N-methylpyrrole-N-methylimidazole hairpin polyamide. Biol Pharm Bull 2004; 27: 468–74
- Dervan PB. Molecular recognition of DNA by small molecules. Bioorg Med Chem 2001; 9: 2215–35
- Kielkopf CL, White S, Szewczyk JW, et al. A structural basis for recognition of A.T and T.A base pairs in the minor groove of B-DNA. Science 1998; 282: 111–5
- Wang CC, Ellervik U, Dervan PB. Expanding the recognition of the minor groove of DNA by incorporation of beta-alanine in hairpin polyamides. Bioorg Med Chem 2001; 9: 653–7
- Ayame H, Saito T, Bando T, Fukuda N, Sugiyama H. Fmoc solid-phase synthesis and its application to pyrrole-imidazole polyamides. Nucleic Acids Res 2003; 3: 67–8s
- Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, et al. Mutations of the VHL tumor suppressor gene in renal carcinoma. Nature Genet 1994; 7: 85–90
- Xia G, Kageyama Y, Hayashi T, Kawakami S, Yoshida M, Kihara K. Regulation of vascular endothelial growth factor transcription by endothelial PAS domain protein 1 (EPSA1) and possible involvement of EPAS1 in the angiogenesis of renal cell carcinoma. Cancer 2001; 91: 1429–36
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia inducible factor 1α is mediated by an O2-dependent degradation domain via an ubiquitin-proteasome pathway. Proc Natl Acad Sci USA 1998; 95: 7978–92
- Kageyama Y, Koshiji M, To KKW, Tian YM, Ratcliffe PJ, Huang LE. Leu-574 of human HIF-1a is a molecular determinant of prolyl hydroxylation. FASEB J 2004; 18: 1028–30
- Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barret JC, Hunag LE. HIF-1α induces cell cycle arrest functionally counteracting Myc. EMBO J 2004; 23: 1949–56
- Shibata T, Akiyama N, Noda M, Sasai K, Hiraoka M. Enhancement of gene expression under hypoxic conditions using fragments of the human vascular endothelial growth factor and the erythropoietin genes. Int J Radiat Oncol Biol Phys 1998; 42: 913–26
- Takahashi R, Bando T, Sugiyama H. Specific alkylation of human telomere repeats by hairpin pyrrole-imidazole polyamide. Bioorg Med Chem 2003; 11: 2503–9
- Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WGJr., Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc Natl Acad Sci USA 2004; 101: 16768–73
- Oyoshi T, Kawakami W, Narita A, Bando T, Sugiyama H. Inhibition of transcription at a coding sequence by alkylating polyamide. J Am Chem Soc 2003; 125: 4752–4
- Shinohara K, Narita A, Oyoshi T, Bando T, Teraoka H, Sugiyama H. Sequence-specific gene silencing in mammalian cells by alkylating pyrrole-imidazole polyamides. J Am Chem Soc 2004; 126: 5113–8
- Linehan WM, Vasselli J, Srinivasan R, Walther MM, Merino M, Choyke P, et al. Genetic basis of cancer of the kidney: disease-specific approaches to therapy. Clin Cancer Res 2004; 10: 6282–9s
- Iwai A, Fujii Y, Kawakami S, Takazawa R, Kageyama Y, Yoshida MA, et al. Down-regulation of vascular endothelial growth factor in renal cell carcinoma cells by glucocorticoids. Mol Cell Endocrinol 2004; 226: 11–7
- Seagroves T, Johnson RS. Two HIFs may be better than one. Cancer Cell 2002; 1: 211–3
